Multiple sequence-alignment-based RNA language model and its application to structural inference
This repository contains codes and pre-trained weight for MSA RNA language model (RNA-MSM) as well as RNA secondary structure and solvent accessibility tasks and corresponding RNA datasets.
RNA-MSM is the first unsupervised MSA RNA language model based on aligned homologous sequences that outputs both embedding and attention map to match different types of downstream tasks.
The resulting RNA-MSM model produced attention maps and embeddings that have direct correlations to RNA secondary structure and solvent accessibility without supervised training. Further supervised training led to predicted secondary structure and solvent accessibility that are significantly more accurate than current state-of-the-art techniques. Unlike many previous studies, we would like to emphasize that we were extremely careful in avoiding over training, a significant problem in applying deep learning to RNA by choosing validation and test sets structurally different from the training set.
Download this repository and create the RNA-MSM environment.
git clone [email protected]:yikunpku/RNA-MSM.git
cd ./RNA-MSM
conda env create -f environment.yml
conda activate RNA-MSM
RNA-MSM model operate on RNA homologous sequences (multiple sequence alignment; MSA), which contains information about conserved properties, co-evolution and functional-species evolutionary relationships (phylogenetics) in the amino acid sequences of constituent RNAs.
The effectiveness of predictions made by the RNA-MSM model is largely dependent on the quantity and quality of MSAs. Therefore, we recommend utilizing our recently developed RNAcmap3 tool to search for homologous sequences of the target RNA sequences to serve as input for the RNA-MSM model.
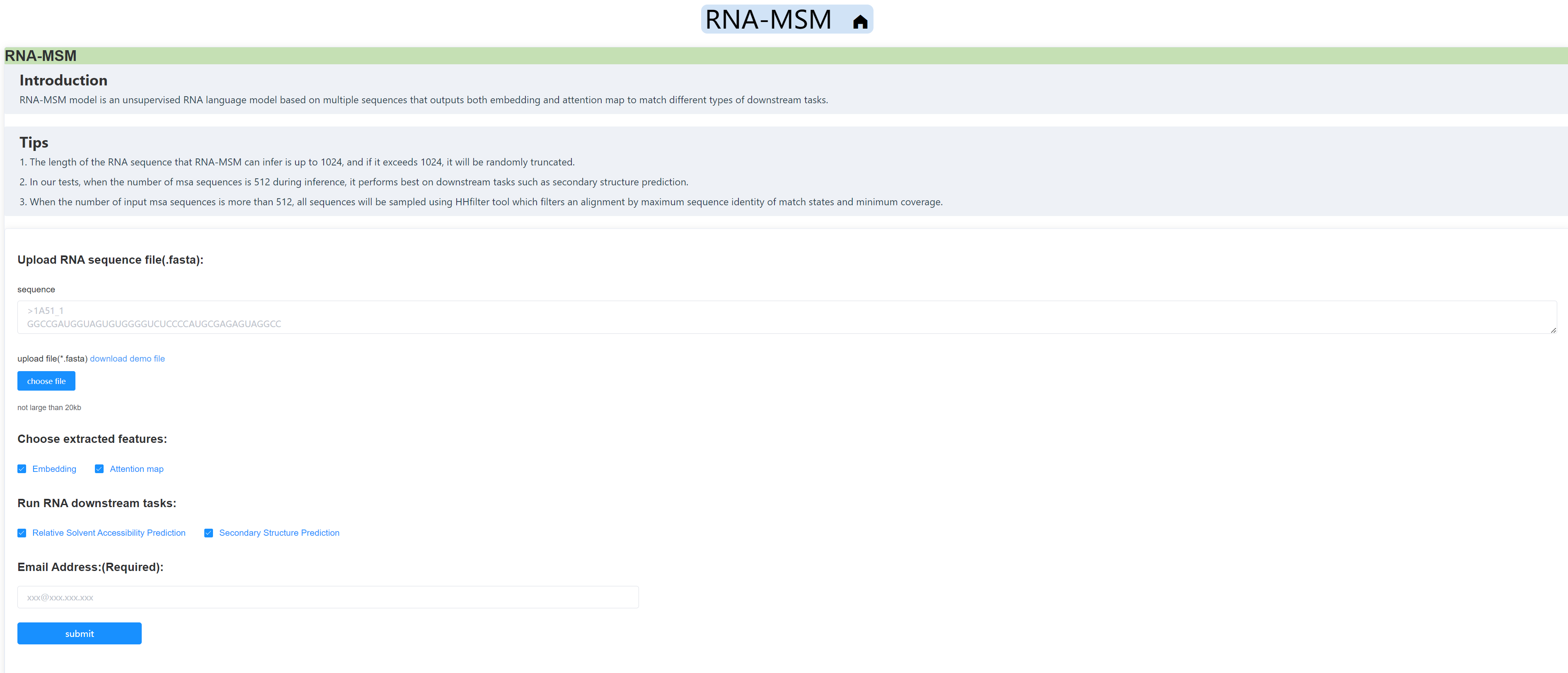
You may also gain entry to our online web server, wherein you can provide the target sequence, and subsequently receive the MSA files and two downstream tasks prediction results located via email.
The input MSA file should be be situated within ./results folder, and its suffix ought to be .a2m_msa2.
The training, validation, and testing datasets used for our downstream tasks are currently available to the public and can be downloaded via this link.
Download pre-trained models from and place the .ckpt files into the ./pretrained folder.
To following command can be used to extract target RNA sequence’s embedding and attention map feature:
python RNA_MSM_Inference.py \
data.root_path=./ \
data.MSA_path=./results \
data.model_path=./pretrained \
data.MSA_list=rna_id.txt
Generated files are saved at data.root_path/data.MSA_path
RNA-MSM model inference results includes 2 files:
-
*_atp.npy: Attention heads weights of the target RNA sequence generated by our RNA-MSM model with dimension (seq_len, seq_len, 120), saved as .npy format. You can apply this embedding feature to your own tasks. -
*_emb.npy: Embedding representation of the target RNA sequence generated by our RNA-MSM model with dimension (seq_len, 768), saved as .npy format. You can apply this embedding feature to your own tasks.
cd ./_downstream_tasks/SS
python predict.py \
--rnaid 2DRB_1 \
--device cpu \
--featdir ./results
In addition, the following arguments need to be specified:
--rnaid :target RNA name, eg: 2DRB_1
--device:inference on GPU or CPU
--featdir: inference output dir
Generated files are saved at data.root_path/data.MSA_path
RNA secondary structure prediction results include 3 files:
*.ct: CT file. The connect format is column based. The first column specified the sequence index, starting at one. Columns 3, 4, and 6 redundantly give sequence indices (plus/minus one). The second column contains the base in one-letter notation. Column 4 specifies the pairing partner of this base if it involved in a base pair. If the base is unpaired, this column is zero.*.bpseq: The structural information in the bpseq format is denoted in three columns. The first column contains the sequence position, starting at one. The second column contains the base in one-letter notation. The third column contains the pairing partner of the base if the base is paired. If the base is unpaired, the third column is zero.*.prob:a 2-dimension matrix that contain the probability of all base-pairs.
cd ./_downstream_tasks/RSA
python predict.py \
python predict.py \
--rnaid 2DRB_1 \
--device cpu \
--featdir ./results
Generated files are saved at data.root_path/data.MSA_path
Solvent accessibility prediction results include 6 files:
*_asa.png: Graph of ASA predicted by ensemble model.*_rsa.png: Graph of RSA predicted by ensemble model.- Results predicted by single model :
model_0is the best single model, other 2 files are remain models。 - Results predicted by ensemble model :
ensembleis the results predicted by ensemble model.
We show the final result directory as follow:
./results
|-- 2DRB_1.a2m_msa2
|-- 2DRB_1_atp.npy
|-- 2DRB_1_emb.npy
|-- RSA_result
| |-- 2DRB_1_asa.png
| |-- 2DRB_1_rsa.png
| |-- ensemble
| | `-- 2DRB_1.txt
| |-- model_0
| | `-- 2DRB_1.txt
| |-- model_1
| | `-- 2DRB_1.txt
| `-- model_2
| `-- 2DRB_1.txt
`-- SS_result
|-- 2DRB_1.bpseq
|-- 2DRB_1.ct
`-- 2DRB_1.prob
We also built a freely accessible web server for using the RNA-MSM models, You may effortlessly submit tasks onto the server and subsequently receive the outcomes via email, without the need to configure the environment or consume any computational resources.
As a preview, take a swift glance at the website:
If you find our work useful in your research or if you use parts of this code please consider citing our paper:
@article{zhang2023multiple,
title={Multiple sequence-alignment-based RNA language model and its application to structural inference},
author={Zhang, Yikun and Lang, Mei and Jiang, Jiuhong and Gao, Zhiqiang and Xu, Fan and Litfin, Thomas and Chen, Ke and Singh, Jaswinder and Huang, Xiansong and Song, Guoli and others},
journal={bioRxiv},
pages={2023--03},
year={2023},
publisher={Cold Spring Harbor Laboratory}
}