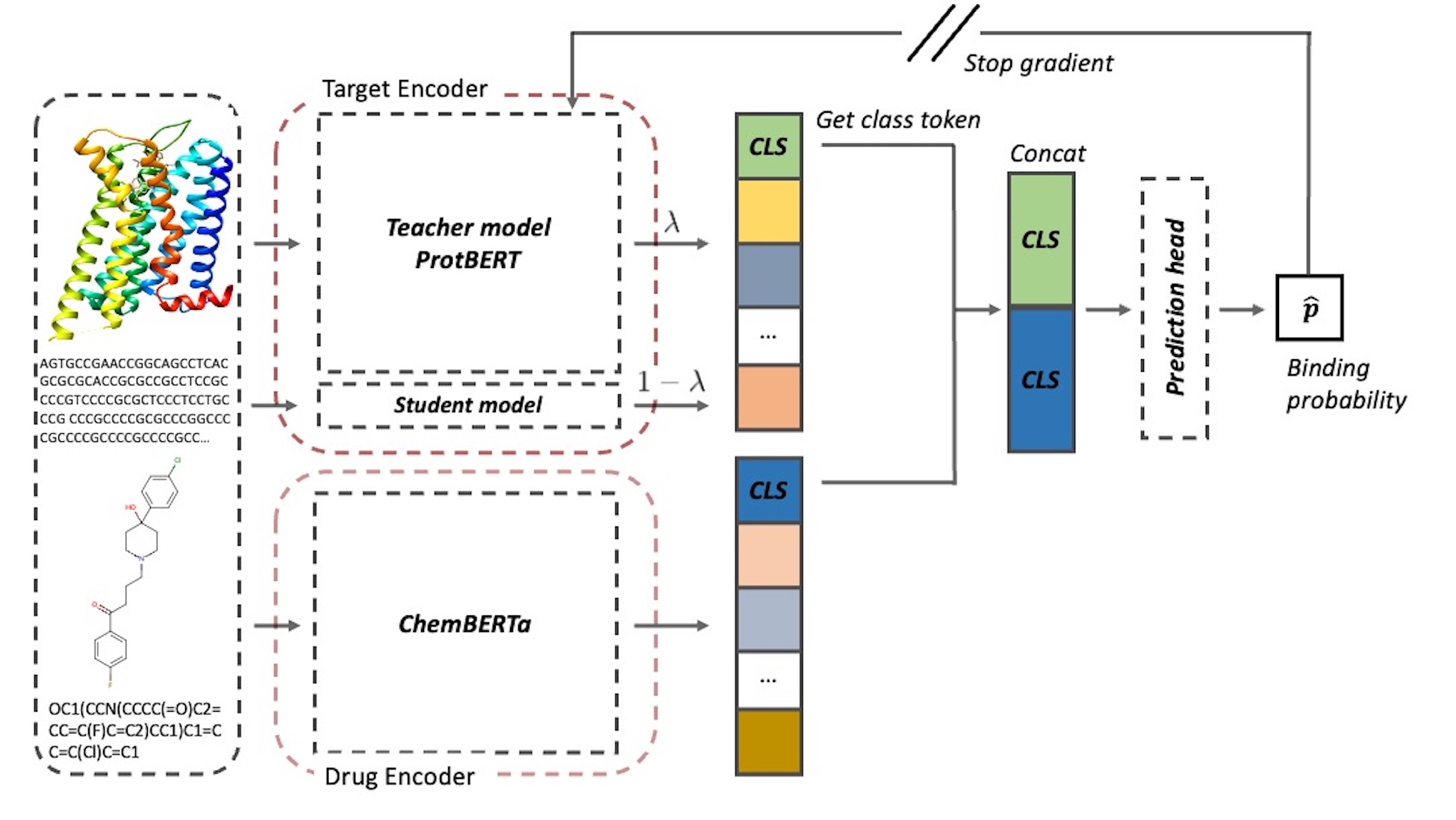
The drug discovery process is demanding and time-consuming, and machine learning-based research is increasingly being proposed to enhance efficiency. A significant challenge in this field is predicting whether a drug molecule structure will interact with a target protein. A recent study attempted to address this challenge by utilizing an encoder that leverages prior knowledge of molecular and protein structures, resulting in notable improvements in the prediction performance of the drug-target interactions task. Nonetheless, the target encoders employed in previous studies exhibit computational complexity that increases quadratically with the input length, thereby limiting their practical utility. To overcome this challenge, we adopt a hint-based learning strategy, to develop a compact and efficient target encoder. With the adaptation parameter, our model could mix the general knowledge and target-oriented knowledge to build features of the protein sequences. This approach yielded considerable performance and enhancements of learning efficiency on three benchmark datasets: BIOSNAP, DAVIS, and BindingDB. Furthermore, our methodology boasts the merit of necessitating only a minimal Video RAM (VRAM) allocation, specifically 7.7GB, during the training phase. This ensures the feasibility of training and inference even with constrained computational resources.
- Drug (molecule) encoder
- Target (protein) encoder
- Fine-tuning of BERT Model to Accurately Predict Drug–Target Interactions
- Fit-Net (hint-based learning)
- Binary classification: DAVIS, Binding DB, and BIOSNAP
- Downloaded from MolTrans
- You can install the required libraries by running
pip install -r requirements.txt - If you encounter any installation errors, please don't hesitate to reach out to us for assistance.
- You can run experiments using config files (under config folder). For example you can launch an experiment by running
python run.py -c config/BindingDB_prot-545_lambda-learnable.yamlorpython run.py --config config/BindingDB_prot-545_lambda-learnable.yaml - Hyperparameters are recorded in config file, therefore, you can modify experiments easily.
Example config file
dataset: BindingDB # There are four options; DAVIS, BindingDB, BIOSNAP, and merged.
device: 0 # GPU device number. If you have a single GPU, the device number is 0.
prot_length:
teacher: 545
student: 545
lambda:
learnable: True
fixed_value: -1
prot_encoder:
hidden_size: 1024
num_hidden_layers: 2
num_attention_heads: 16
intermediate_size: 4096
hidden_act: "gelu"
training_config:
batch_size: 32
num_workers: 16
epochs: 50
hidden_dim: 1024
learning_rate: 0.0001
The archived version of the source code and data were stored in Zenodo (7/Sep/2023).