🚧 This repository is still under construction. 🚧
Please feel free to explore and contribute, but note that there may be frequent changes.
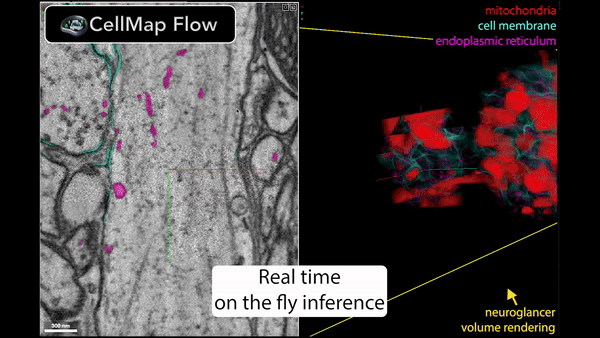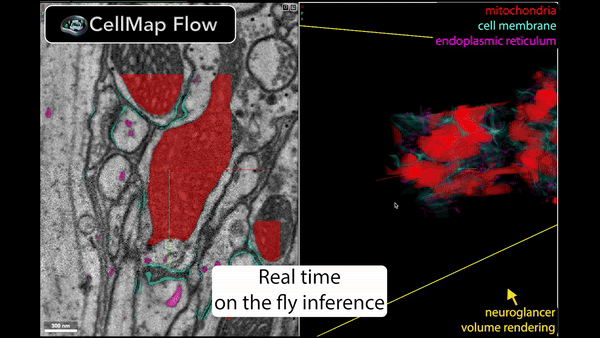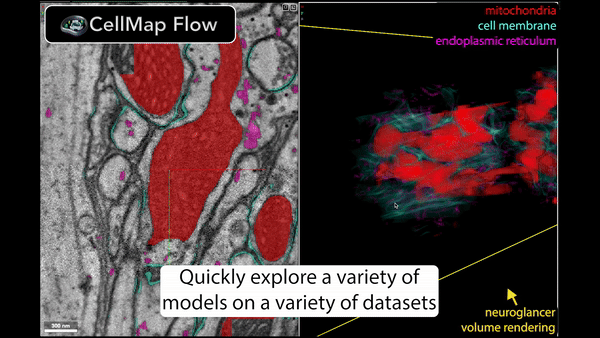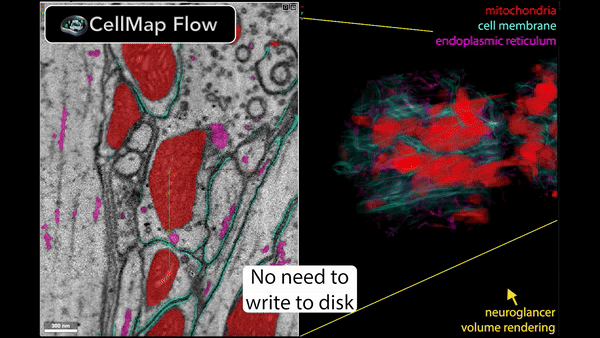
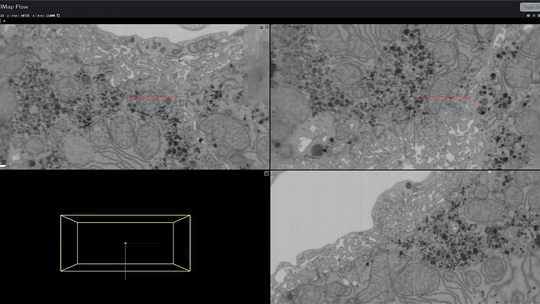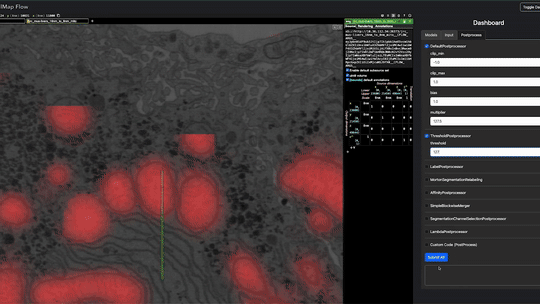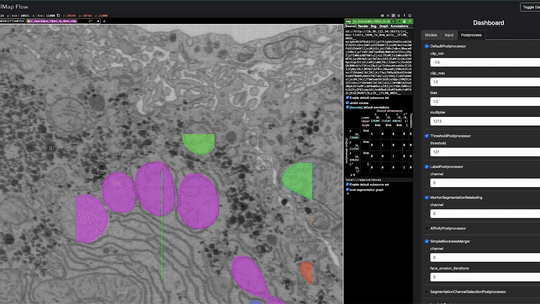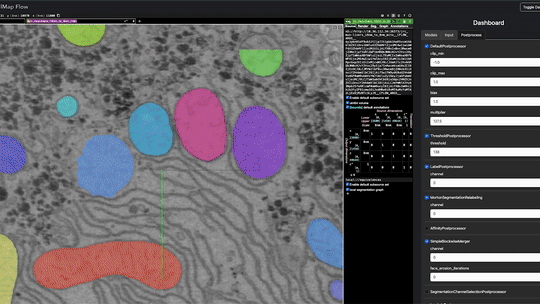
Real-time inference is performed using Torch/Tensorflow, Dacapo, and bioimage models on local data or any cloud-hosted data.
🚀 Speed up your data processing from months to minutes!
To install CellMapFlow, you can use pip:
pip install cellmap-flowNote that the basic installation does not include DaCapo and BioImage.io core dependencies. To install CellMapFlow with DaCapo support, use the following command:
pip install cellmap-flow[dacapo]To install CellMapFlow with BioImage.io support, use the following command:
pip install cellmap-flow[bioimage]To install CellMapFlow with both DaCapo and BioImage.io support, use the following command:
pip install cellmap-flow[dacapo,bioimage]$ cellmap_flow
Usage: cellmap_flow [OPTIONS] COMMAND [ARGS]...
Examples:
To use Dacapo run the following commands:
cellmap_flow dacapo -r my_run -i iteration -d data_path
To use custom script
cellmap_flow script -s script_path -d data_path
To use bioimage-io model
cellmap_flow bioimage -m model_path -d data_path
Commands:
bioimage Run the CellMapFlow server with a bioimage-io model.
dacapo Run the CellMapFlow server with a DaCapo model.
script Run the CellMapFlow server with a custom script.Currently available:
This enables using any model by providing a script e.g. example/model_spec.py e.g.
cellmap_flow script -s /groups/cellmap/cellmap/zouinkhim/cellmap-flow/example/model_spec.py -d /nrs/cellmap/data/jrc_mus-cerebellum-1/jrc_mus-cerebellum-1.zarr/recon-1/em/fibsem-uint8/s0 Define these variables in your script (cellmap_flow script -s path/to/your_script.py):
- model: The PyTorch model to be used for inference.
- input_size: The voxel shape of the data to be input to the PyTorch model.
- output_size: The voxel shape of the data in output by the PyTorch model.
- input_voxel_size: The voxel size of the data input to the model.
- output_voxel_size: The voxel size of the data output by the model.
- output_channels: The number of channels in the output of the model.
- process_chunk (optional): (Optional) A function that takes an ImageDataInterface and an ROI and returns the data to be display. This can be used to run a TensorFlow model or do other custom data processing.
which enable inference using a Dacapo model by providing the run name and iteration number e.g.
cellmap_flow dacapo -r 20241204_finetune_mito_affs_task_datasplit_v3_u21_kidney_mito_default_cache_8_1 -i 700000 -d /nrs/cellmap/data/jrc_ut21-1413-003/jrc_ut21-1413-003.zarr/recon-1/em/fibsem-uint8/s0still in development
To run TensorFlow models, we suggest installing TensorFlow via conda: conda install tensorflow-gpu==2.16.1
cellmap_flow_multiple --script -s /groups/cellmap/cellmap/zouinkhim/cellmap-flow/example/model_spec.py -n script_base --dacapo -r 20241204_finetune_mito_affs_task_datasplit_v3_u21_kidney_mito_default_cache_8_1 -i 700000 -n using_dacapo -d /nrs/cellmap/data/jrc_ut21-1413-003/jrc_ut21-1413-003.zarr/recon-1/em/fibsem-uint8/s0