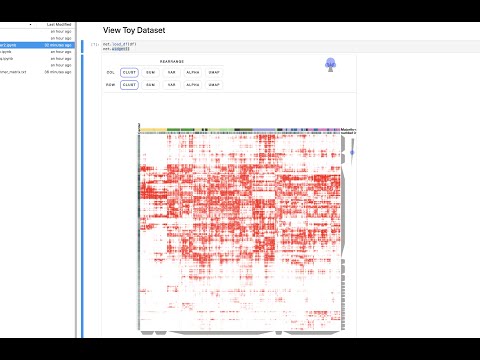
Clustergrammer2 is an interactive heatmap Jupyter widget built using the widget-ts-cookiecutter library and the WebGL library regl. Clustergrammer2 is built to help researchers interactively explore single cell data (e.g. scRNA-seq). Please see Case Studies and Tutorials for examples.
The above notebook shows how Clustergrammer2 can be used to load a small dataset and visualize a large random DataFrame. By running the notebook on MyBinder using Jupyter Lab it can also be used to visualize a user uploaded dataset. Please see the video tutorial above for more information.
For additional examples and tutorials please see:
- Case Studies and Tutorials
- Clustergrammer2-Notebooks GitHub repository
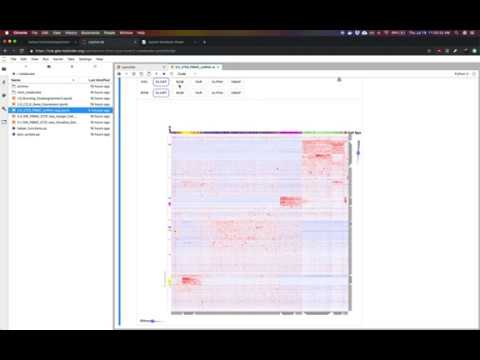
Single cell RNA-seq (scRNA-seq) is a powerful method to interrogate gene expression across thousands of single cells. This method produces thousands of measurements (single cells) across thousands of dimensions (genes). This notebook uses Clustergrammer2 to interactively explore an example dataset measuring the gene expression of 2,700 PBMCs obtained from 10X Genomics. Bulk gene expression signatures of cell types from CIBERSORT were used to obtain a tentative cell type for each cell. Please see the video tutorial above for more information.
Please see the project Roadmap for information on where the project is heading.
A typical installation requires the following commands to be run:
pip install clustergrammer2
jupyter nbextension install --py --sys-prefix clustergrammer2
jupyter nbextension enable --py --sys-prefix clustergrammer2Optionally: --sys-prefix|--user|--system
Or, if you use jupyterlab:
pip install clustergrammer2
jupyter labextension install @jupyter-widgets/jupyterlab-managerjupyter nbconvert --to html notebook.ipynbDuring development run npm run watch for real time updates. When releasing a new version, first confirm that latest version of the front and back ends are working locally (check console logs, etc).
Update the versions in the following files
- package.json
- widget.ts
- _version.py
- example.py
- requirements.txt
- _frontend.py
Run the following commands to build the JavaScript bundle:
npm run build
npm run build:nbextension
npm run build:labextensionPublish to npm using
npm publishThese instructions are based on the release instructions from the jupyter-widgets/widget-ts-cookiecutterREADME.
Next, bundle the python package using (optinally delete the old versions under dist)
python setup.py sdist bdist_wheelThen, upload the PYPI:
twine upload dist/*After releasing a new version several things need to be checked to ensure proper widget functioning.
- https://www.npmjs.com/package/clustergrammer2 (can be slow to update)
- https://pypi.org/project/clustergrammer2/
- Run
jupyter nbconvert --to html introduction_nb.ipynb(from the examples directory) to generate a static HTML and check that the latest widget is working (uses unpkg.com, like https://unpkg.com/[email protected]/dist/index.js) - Check NBViewer using link to notebook on GitHub (e.g. https://nbviewer.jupyter.org/github/ismms-himc/clustergrammer2/blob/master/examples/introduction_nb.ipynb?flush_cache=true). Make sure to use the querystring
flush_cache=Truein the URL to ensure that NBVIewer re-runs nbconvert.
- Check MyBinder
Use this in the where section to restrict search
clustergrammer2/*.py,-*/build/*
For issues and concerns please use the issue tracker or gitter discussion room.