⚠️ cytodatagen takes a very naive approach to generate cytometry data. You might want to checkout FlowCyPy instead! FlowCyPy attempts to generate realistic SSC and FSC signals by regarding the fluiddynamics, optics and electronics of a flow cytometer. However, at the time of writing, FlowCyPy didn't yet support generating flourescence signals.
Generate synthetic cytometry data for classification tasks
This package provides tools to generate synthetic flow cytometry/CyTOF data for classification tasks.
Supported formats are fcs and h5ad.
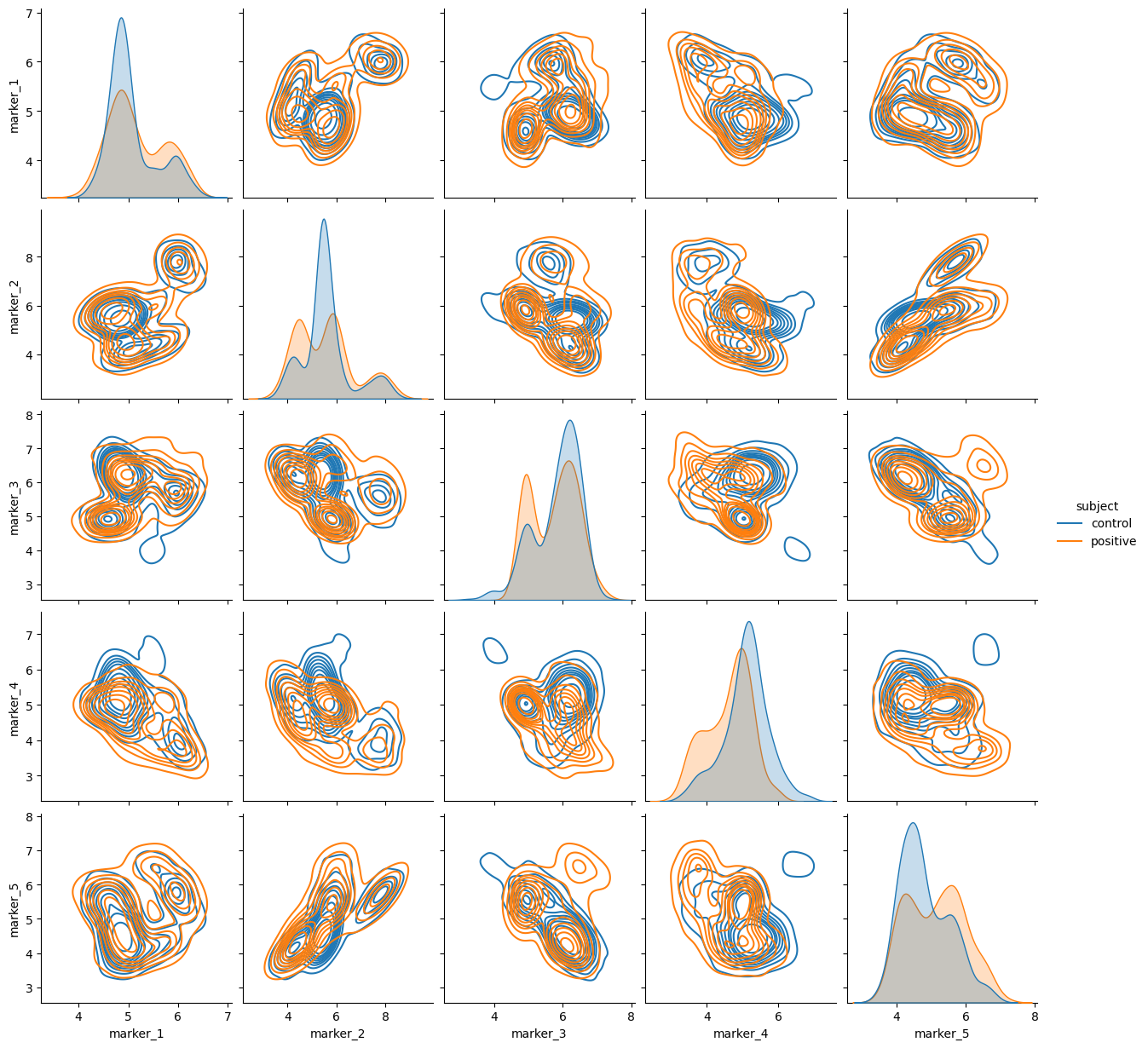
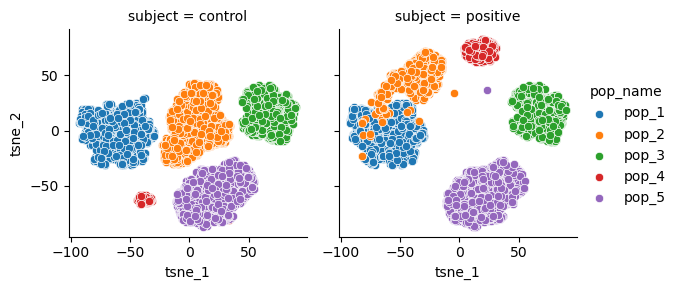
The synthetic data generated with cytodatagen assumes that subjects differ either in:
- distribution shifts of populations
- example: T-cells have a higher marker value in postive subjects
- cell type composition
- example: positive subjects have a higher T-cell count
# install via pip
pip install cytodatagen
# or install from source
pip install -e .
# install with extras for jupyter notebooks
pip install -e .[notebook]The package provides a CLI:
# display help message
python -m cytodatagen.cli.subjects --help
# generate subjects.json configuration file and adjust it later
python -m cytodatagen.cli.subjects config/cli/subjects/config.json -o artifacts/subjects.json --seed 19
# display help message
python -m cytodatagen.cli.data --help
# generate data from command line
python -m cytodatagen.cli.data -o artifacts/cytodata --format fcs
python -m cytodatagen.cli.data -o artifacts/demo --format h5ad --subjects config/cli/data/subjects.json --transforms config/cli/data/transforms.json --seed 19Look at the scripts scripts/make_subjects.sh and scripts/make_data.sh for more examples.
- FlowCyPy
- Yang et al.: Cytomulate: accurate and efficient simulation of CyTOF data (2023)
- Cheung et al.: Systematic Design, Generation, and Application of Synthetic Datasets for Flow Cytometry (2022)
Thanks to the FlowKit and anndata team.
This project has been set up using PyScaffold 4.6. For details and usage information on PyScaffold see https://pyscaffold.org/.