Full documentation: here
Intro to MELD Graph and installation videos: here
Graph based FCD lesion segmentation for the MELD project.
This package is a pipeline to segment FCD-lesions from MRI scans.
SIGN UP TO THE MELD GRAPH MAILING LIST:
We request that all MELD Graph users sign up to the mailing list. If you are using MELD Graph, please send an email to [email protected] with the subject 'Request to be added to the MELD Graph mailing list' and provide use with your name and institute. This will ensure that we can update you about bug fixs and new releases.
EXISTING USERS: PLEASE UPDATE TO VERSION V2.2.2: We have released MELD Graph V2.2.2 which fixes a couple of issues found by users. For more information about the release please see MELD Graph V2.2.2. To update your code please follow the guidelines Updating MELD Graph to V2.2.2 from our FAQ.
Code Authors : Mathilde Ripart, Hannah Spitzer, Sophie Adler, Konrad Wagstyl
This package is intended to be used as a research tool to segment FCD lesions in patients with focal epilepsy where a FCD is suspected. It can be run on 1.5T or 3T MRI data. A 3D T1 is required and it is optional but advised to include the 3D FLAIR.
It is not appropriate to use this algorithm on patients with:
- suspected hippocampal sclerosis
- hypothalamic hamartoma
- periventricular nodular heterotopia
- other focal epilepsy pathologies
- previous resection cavities
Harmonisation - MRI data from different MRI scanners looks subtly different. This means that feature measurements, e.g. cortical thickness measurements, differ depending on which MRI scanner a patient was scanned on. We harmonise features (using NeuroCombat) to adust for site based differences. We advise new users to harmonise data from their MRI scanner to the MELD graph dataset. Please follow the guidelines to harmonise the data from your site. Note: the model will still produce predictions on new, unharmonised subjects but the number of false positive predictions is higher if the data is not harmonised.
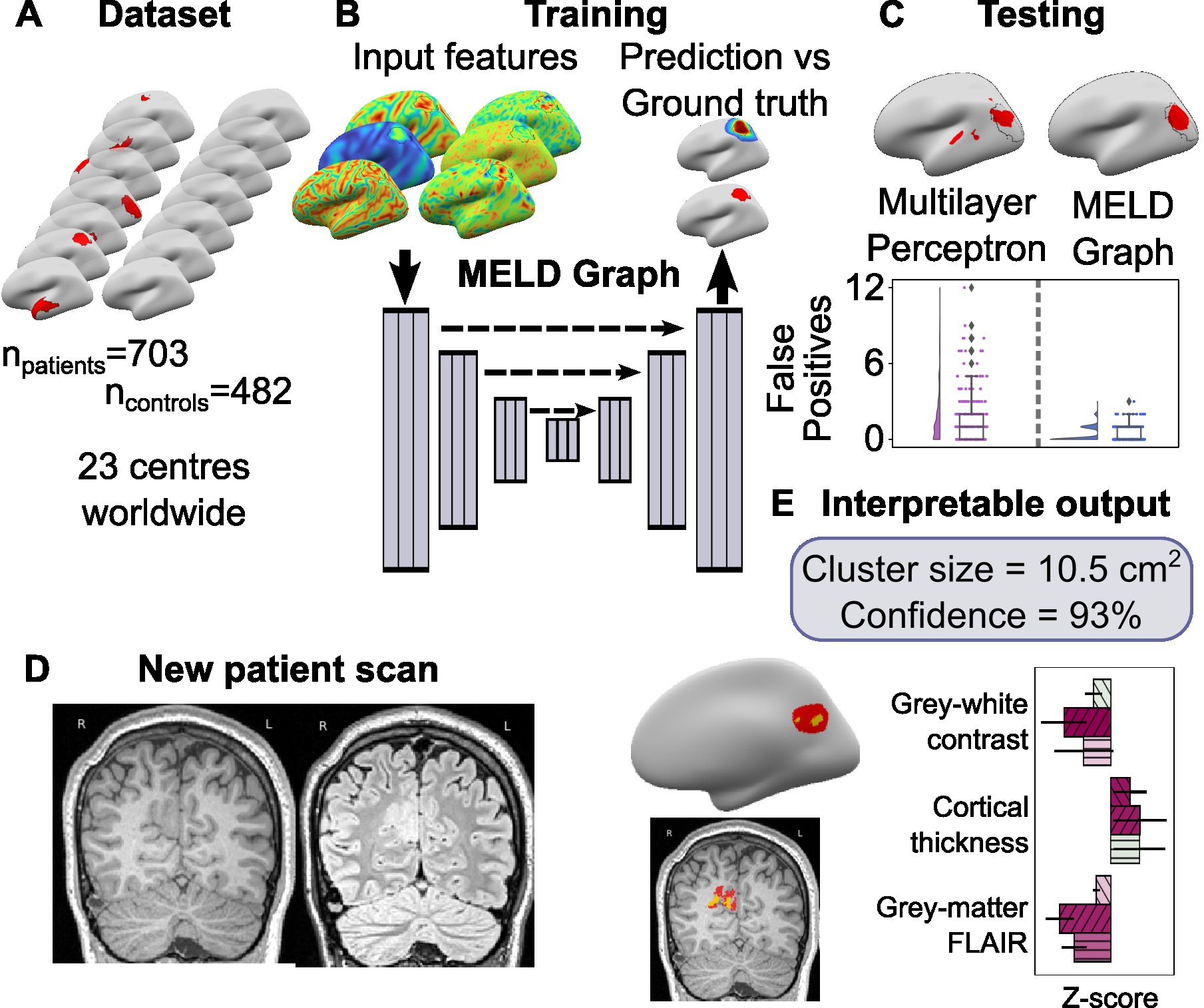
This package also contains code for training and evaluating graph-based U-net lesion segmentation models operating on icosphere meshes.
In addition to lesion segmentation, the model also contain auxiliary distance regression and hemisphere classification losses.
For more information on how the algorithm was developed and expected performance - check our papers:
- Ripart et al.,2025 JAMA Neurology - Detection of epileptogenic focal cortical dysplasia using graph neural networks: a MELD study
- Spitzer, Ripart et al., 2022 Brain - the original MELD FCD pipeline and dataset
- Spitzer et al., 2023 MICCAI - the updated graph-based model architecture
The MELD surface-based graph FCD detection algorithm is intended for research purposes only and has not been reviewed or approved by the Medicines and Healthcare products Regulatory Agency (MHRA), European Medicine Agency (EMA) or by any other agency. Any clinical application of the software is at the sole risk of the party engaged in such application. There is no warranty of any kind that the software will produce useful results in any way. Use of the software is at the recipient's own risk.
You can install and use the MELD FCD prediction pipeline with :
- docker container recommended for easy installation of the pipeline as all the prerequisite packages are already embedded into the container. Note: Dockers are not working on High Performance Computing (HCP) systems
- native installation recommended for Mac and users that want to modify the code and/or use the code to train/test their own classifier.
- singularity container enables to run a container on High Performance Computing (HCP) systems.
IMPORTANT NOTE: The installations listed above are not supported on Virtual Machines. Please install MELD Graph on full Linux, Windows or MAC computers
YouTube tutorials available for the docker installation and native installation
Once installed you will be able to use the MELD FCD prediction pipeline on your data following the steps:
- Prepare your data : guidelines
- Compute the harmonisation parameters : guidelines (OPTIONAL but highly recommended)
- Run the prediction pipeline: guidelines
- Interpret the results: guidelines
YouTube tutorials available to run the harmonisation step, to run the prediction pipeline and to interpret the pipeline results
FAQs If you have a question or if you are running into issues at any stage (installation/use/interpretation), have a look at our FAQs page as we may have already have a solution.
What is the harmonisation process ?
Scanners can induce a bias in the MRI data. The MELD pipeline recommends adjusting for these scanners differences by running a preliminary harmonisation step to compute the harmonisation parameters for that specific scanner. Note: this step needs to be run only once, and requires data from at least 20 subjects acquired on the same scanner and demographic information (e.g age and sex). See harmonisation instructions for more details.
Note: The MELD pipeline can also be run without harmonisation, with a small drop in performance.
With the native installation of the MELD classifier you can reproduce the figures from our paper and train/evaluate your own models. For more details, check out the guides linked below:
If you'd like to contribute to this code base, have a look at our contribution guide
We would like to thank
- the MELD consortium for providing the data to train this classifier and their expertise to build this pipeline.\
- Lennart Walger and Andrew Chen, for their help testing and improving the MELD pipeline to v1.1.0. \
- Ulysses Popple for his help building the docs and dockers.
- Cornelius Kronlage highlighting issues in v2.2.1 and suggesting solutions in v2.2.2
Contact the MELD team at [email protected]
Please note that we are a small team and only have one day a week dedicated to the support of the MELD tools (MELD Graph and AID-HS). We will answer your emails as soon as we can!