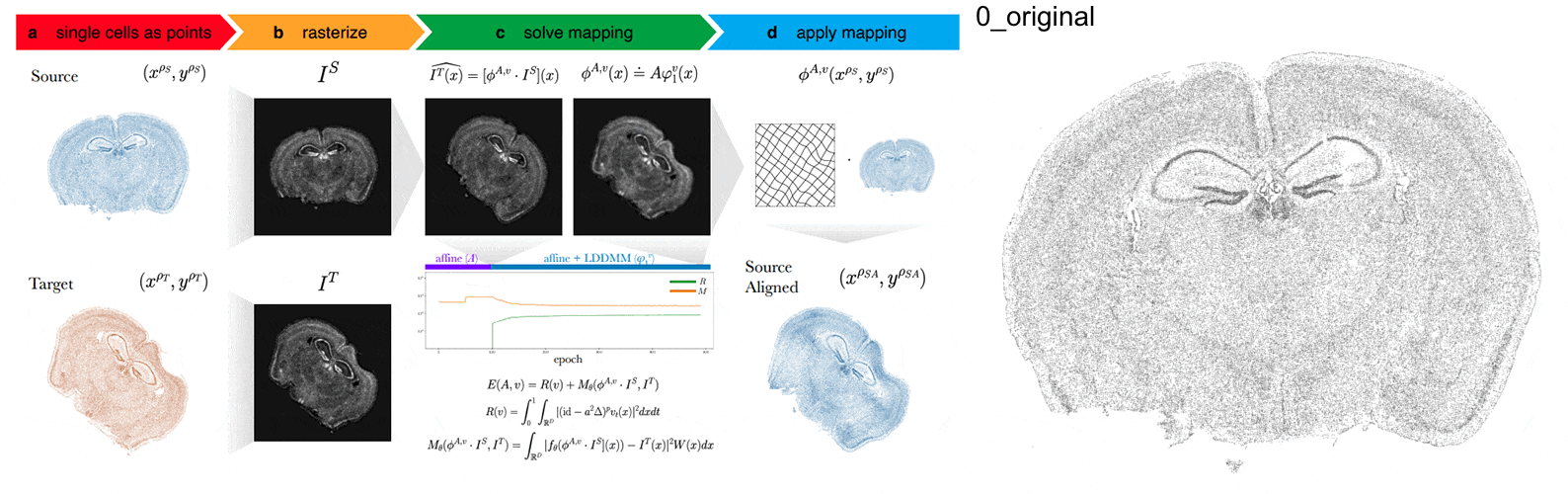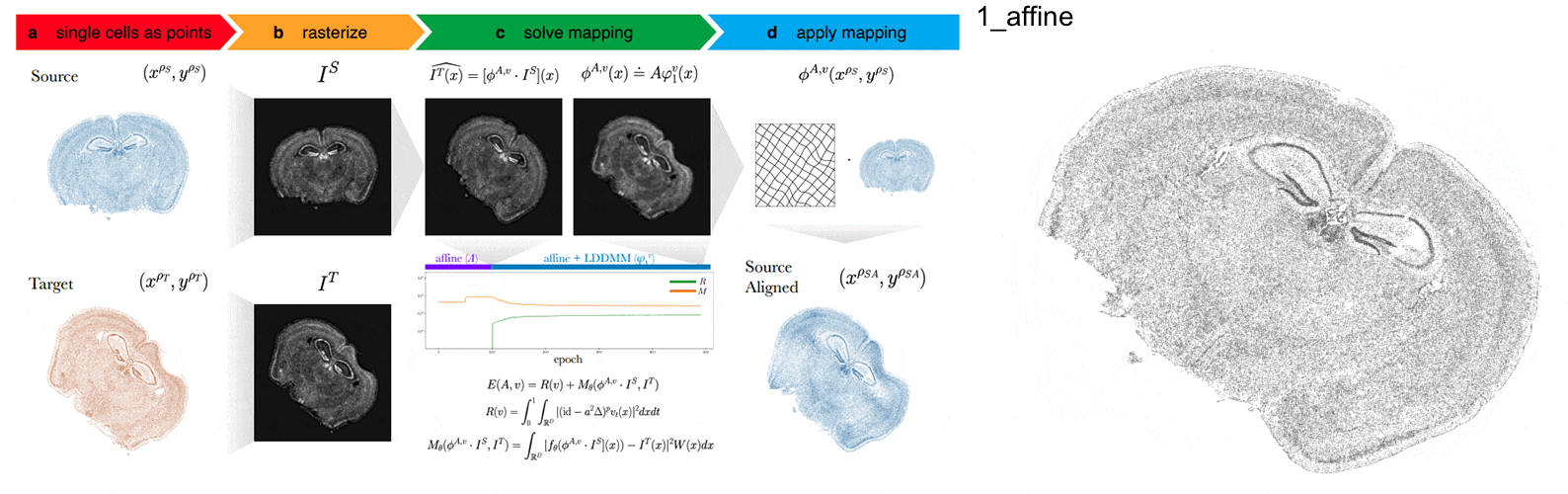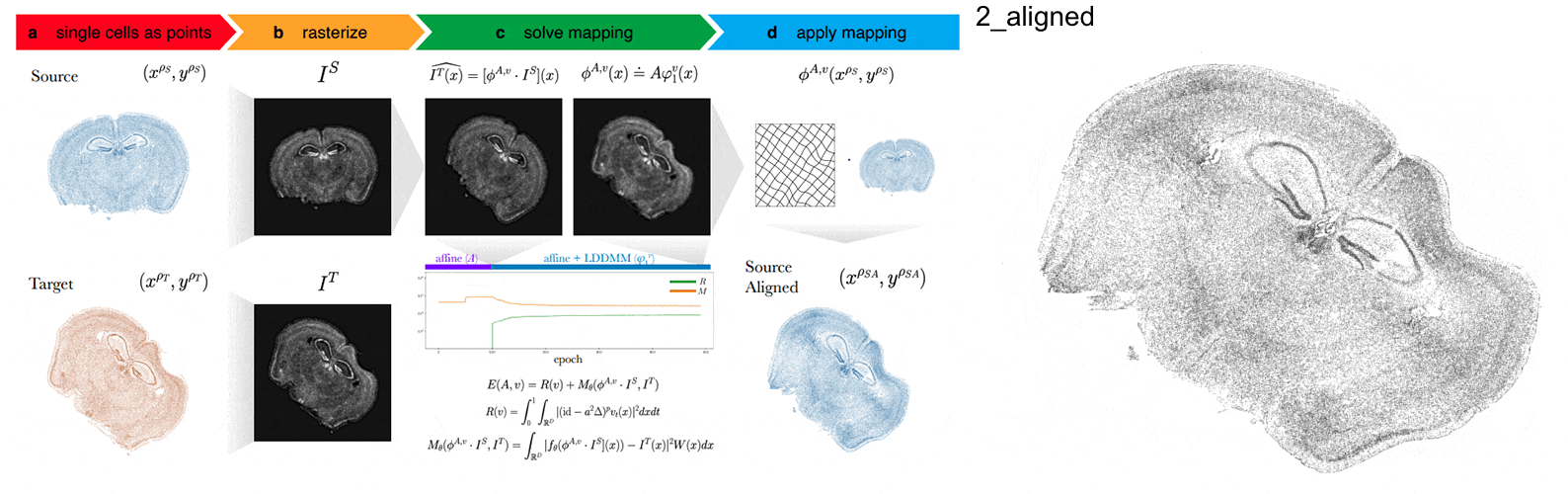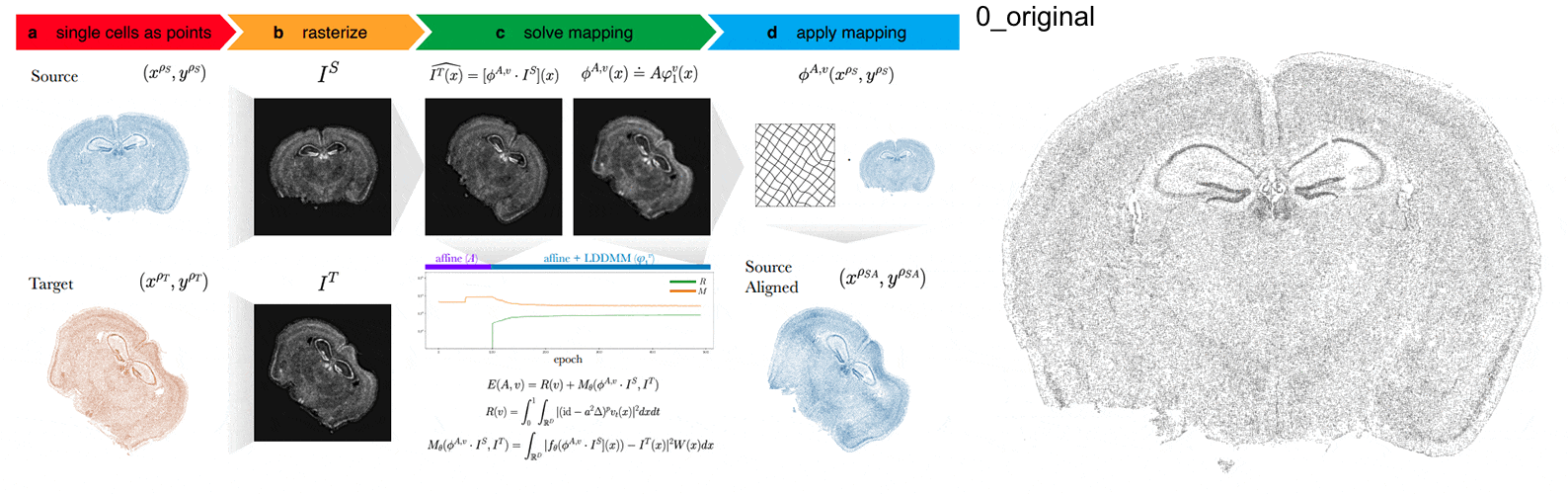
STalign aligns spatial transcriptomics (ST) tissue sections to each other and to 3D atlases like the Allen Brain Atlas using algorithms that build upon diffeomorphic metric mapping.
More information regarding the overall approach, methods and validations can be found in our publication: STalign: Alignment of spatial transcriptomics data using diffeomorphic metric mapping Kalen Clifton*, Manjari Anant*, Gohta Aihara, Lyla Atta, Osagie K Aimiuwu, Justus M Kebschull, Michael I Miller, Daniel Tward^, Jean Fan^
STalign enables:
- alignment of single-cell resolution ST datasets within technologies
- alignment single-cell resolution ST datasets to histology
- alignment of ST datasets across technologies
- alignment of ST datasets to a 3D common coordinate framework
This installation method is intended for users who sets up a Python environment without pipenv.
pip install --upgrade "git+https://github.com/JEFworks-Lab/STalign.git"
All dependencies will be installed into your selected environment with the above command. Dependencies can be found in the requirements.txt file.
This installation method is intended for users who sets up a Python environment with pipenv. pipenv allows users to create and activate a virtual environment with all dependencies within the Python project. For more information and installation instructions for pipenv, see https://pipenv.pypa.io/en/latest/.
Fork and git clone the STalign github repository.
From the base directory of your local STalign git repo, create a Pipfile.lock file from Pipfile using:
pipenv install requests
NOTE: Since
Pipfile.lockis platform-dependent and different across operating systems, do not commitPipfile.lockto the git repo if contributing toSTalignor collaborating with other people.
Activate the virtual environment using:
pipenv shell
Deactivate the virtual environment using:
exit
To import STalign into your Python script, use:
from STalign import STalignTo use this tool, you will need provide the following information:
- Source: Arrays of x and y positions of cells
- Target: Arrays of x and y positions of cells
- Source: Arrays of x and y positions of cells from single-cell resolution ST data
- Target: Registered H&E image from spot-resolution ST data
- Source: (Default: Adult mouse Allen Brain Altas CCFv3) 3D Matrix with voxels corresponding to (1) cell intensity and (2) annotated tissue regions
- Target: Arrays of x and y positions of cells
To use STalign, please refer to our tutorials with usage examples.