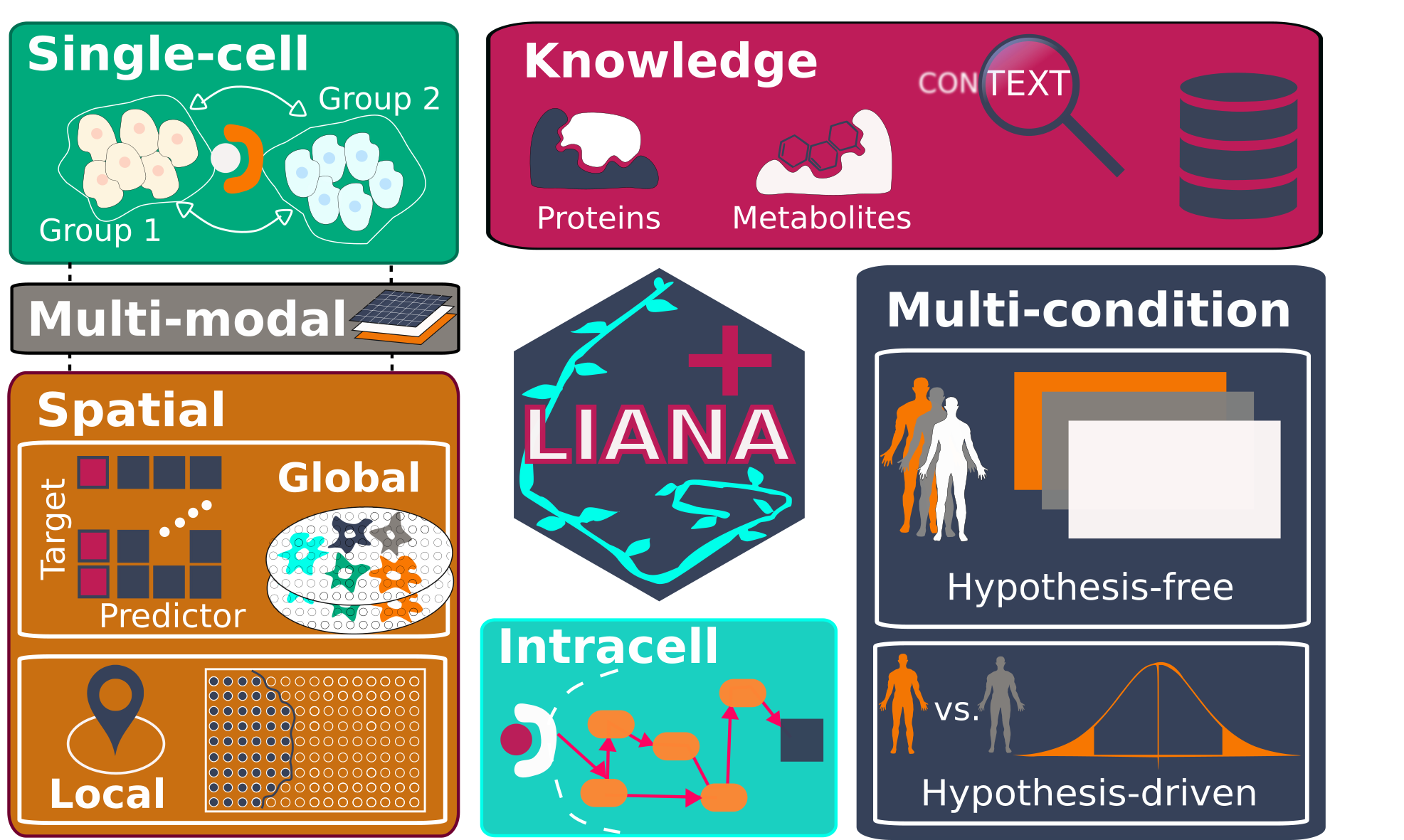
LIANA+ is a scalable framework that integrates and extends existing methods and knowledge to study cell-cell communication in single-cell, spatially-resolved, and multi-modal omics data. It is part of the scverse ecosystem, and relies on AnnData & MuData objects as input.
We welcome suggestions, ideas, and contributions! Please use do not hesitate to contact us, or use the issues or the LIANA+ Development project to make suggestions.
- LIANA's basic tutorial in dissociated single-cell data
-
Learn spatially-informed relationships with MISTy across (multi-) views.
-
Estimate local spatially-informed bivariate metrics. This tutorial shows how to estimate local spatially-informed bivariate metrics, such as the spatially-informed Pearson correlation coefficient or Cosine similarity.
-
Hypothesis-testing for CCC with PyDeSeq2 that also shows the inference of causal intracellular signalling networks, downstream of CCC events.
-
Multicellular programmes with MOFA. Using MOFA to obtain coordinates gene expression programmes across samples and conditions, as done in Ramirez et al., 2023.
-
LIANA with MOFA. Using MOFA to infer intercellular communication programmes across samples and conditions, as initially proposed by Tensor-cell2cell.
-
LIANA with Tensor-cell2cell to extract intercellular communication programmes across samples and conditions. Extensive tutorials combining LIANA & Tensor-cell2cell are available here.
- We also refer users to the Cell-cell communication chapter in the best-practices guide from Theis lab. There we provide an overview of the common limitations and assumptions in CCC inference from (dissociated single-cell) transcriptomics data.
For further information please check LIANA's API documentation.
Dimitrov D., Schäfer P.S.L, Farr E., Rodriguez Mier P., Lobentanzer S., Dugourd A., Tanevski J., Ramirez Flores R.O. and Saez-Rodriguez J. 2023 LIANA+: an all-in-one cell-cell communication framework. BioRxiv. https://www.biorxiv.org/content/10.1101/2023.08.19.553863v1
Dimitrov, D., Türei, D., Garrido-Rodriguez M., Burmedi P.L., Nagai, J.S., Boys, C., Flores, R.O.R., Kim, H., Szalai, B., Costa, I.G., Valdeolivas, A., Dugourd, A. and Saez-Rodriguez, J. Comparison of methods and resources for cell-cell communication inference from single-cell RNA-Seq data. Nat Commun 13, 3224 (2022). https://doi.org/10.1038/s41467-022-30755-0
Similarly, please consider citing any of the methods and/or resources implemented in liana, that were particularly relevant for your research!