-
Notifications
You must be signed in to change notification settings - Fork 1
/
CBB-Retreat-2015.Rpres
669 lines (509 loc) · 22.1 KB
/
CBB-Retreat-2015.Rpres
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
436
437
438
439
440
441
442
443
444
445
446
447
448
449
450
451
452
453
454
455
456
457
458
459
460
461
462
463
464
465
466
467
468
469
470
471
472
473
474
475
476
477
478
479
480
481
482
483
484
485
486
487
488
489
490
491
492
493
494
495
496
497
498
499
500
501
502
503
504
505
506
507
508
509
510
511
512
513
514
515
516
517
518
519
520
521
522
523
524
525
526
527
528
529
530
531
532
533
534
535
536
537
538
539
540
541
542
543
544
545
546
547
548
549
550
551
552
553
554
555
556
557
558
559
560
561
562
563
564
565
566
567
568
569
570
571
572
573
574
575
576
577
578
579
580
581
582
583
584
585
586
587
588
589
590
591
592
593
594
595
596
597
598
599
600
601
602
603
604
605
606
607
608
609
610
611
612
613
614
615
616
617
618
619
620
621
622
623
624
625
626
627
628
629
630
631
632
633
634
635
636
637
638
639
640
641
642
643
644
645
646
647
648
649
650
651
652
653
654
655
656
657
658
659
660
661
662
663
664
665
666
667
668
669
Reproducible Computational Science - Some good, better, and best practices
=====
font-family: 'Helvetica'
width: 1024
height: 768
<p style="text-align: center;">
A two day workshop in two hours (well, not quite)
</p>
<p style="text-align: center;">
Hilmar Lapp, Duke University
</p>
<p style="font-size: smaller; text-align: center">CBB Retreat, September 2015
<br/>
<!-- public domain dedication copied from CC -->
<a rel="license"
href="http://creativecommons.org/publicdomain/zero/1.0/">
<img src="http://i.creativecommons.org/p/zero/1.0/88x31.png" style="border-style: none;" alt="CC0" />
</a>
</p>
Reproducibility crisis
=====
* Only 6 of 56 landmark oncology papers confirmed
* 43 of 67 drug target validation studies failed to reproduce
* Effect size overestimation is common
***
[](http://www.nature.com/nature/focus/reproducibility/index.html)
Reproducibility matters
=====
incremental: true
Lack of reproducibility in science causes significant issues
* For science as an enterprise
* For other researchers in the community
* _For public policy_
Science retracts gay marriage paper
=====
* Science retracted (without lead author's consent) a study of how
canvassers can sway people's opinions about gay marriage
* Original survey data was not made available for independent
reproduction of results (and survey incentives misrepresented, and
sponsorship statement false)
* Two Berkeley grad students attempted to replicate the study and
discovered that the data must have been faked.
Source:
http://news.sciencemag.org/policy/2015/05/science-retracts-gay-marriage-paper-without-lead-author-s-consent
Reproducibility matters
=====
Lack of reproducibility in science causes significant issues
* For science as an enterprise
* For other researchers in the community
* For public policy
* _For patients_
Seizure study retracted after authors realize data got "terribly mixed"
=====
From the authors of **Low Dose Lidocaine for Refractory Seizures in
Preterm Neonates** ([doi:10.1007/s12098-010-0331-7](https://doi.org/10.1007/s12098-010-0331-7):
> The article has been retracted at the request of the authors. After carefully re-examining the data presented in the article, they identified that data of two different hospitals got terribly mixed. The published results cannot be reproduced in accordance with scientific and clinical correctness.
Source: [Retraction Watch](http://retractionwatch.com/2013/02/01/seizure-study-retracted-after-authors-realize-data-got-terribly-mixed/)
Reproducibility matters
=====
Lack of reproducibility in science causes significant issues
* For science as an enterprise
* For other researchers in the community
* For policy making
* For patients
* _For oneself as a researcher_
Reproducibility = Accelerating science, including your own
=====
<p/>
[](https://plus.google.com/+BrunoOliveira/posts/MGxauXypb1Y)
<small>Bruno Oliviera</small>
***
<p/>
* Research that is difficult to reproduce impedes your future self, and your lab.
* More reproducible research is faster to report, in particular when research is dynamic. (Think data, tools, parameters, etc.)
* More reproducible research is also faster to resume or to build on by others.
<p class="highlight" style="margin-top: 10px;">Reproducible science accelerates scientific progress.</p>
Computational research: codified methods yet challenging to reproduce
=====
* _Dependency hell_: Most software has recursive and differing dependencies. Any one can fail to install, conflict with those of others, and their exact versions can affect the results.
* _Documentation gaps_: Code can easily be very difficult to understand if not documented. Documentation gaps and errors may be harmless for experts, but are often fatal for "method novices".
* _Unpredictable evolution_: Scientific software evolves constantly and often in drastic rather than incremental ways. As a result, results, algorithms, and parameters can change in unpredictable ways, and can render code to fail after it worked only a short while months ago.
See [an experiment on reproducing reproducible computational research](https://storify.com/hlapp/reproducibility-repeatability-bigthink)
Overcoming the training gap
=====

[](https://github.com/Reproducible-Science-Curriculum)
***
* Curriculum development hackathon held December 11-14, 2014, at NESCent
* 21 participants comprising statisticians, biologists,
bioinformaticians, open-science activists, programmers, graduate
students, postdocs, untenured and tenured faculty
* Outcome: open source, reusable curriculum for a two-day workshop on reproducibility for computational research
Held 3 workshops so far
=====
* Inaugural workshop May 14-15, 2015 at Duke in Durham, NC
* June 1-2, 2015 at iDigBio in Gainesville, FL
* September 24-25, 2015, at the Duke Marine Lab in Beaufort, NC
* Have made incremental improvements to curriculum and material in between workshops
***

Current Syllabus
=====
**Day 1**
* Motivation of and introduction to Reproducible Research
* Best practices for file naming and file organization
* Best practices for tabular data
* Literate programming and executable documentation of data modification
***
**Day 2**
* Version control and Git
* Why automate?
* Transforming repetitive R script code into R functions
* Automated testing and integration testing
* Sharing, publishing, and archiving for data and code
Exercise: Motivating reproducibility
=====
type: titleonly
Exercise: Motivating reproducibility
=====
type: prompt
<br/><br/>
This is a two-part exercise:
**Part 1:** Analyze + document
**Part 2:** Swap + discuss
Part 1: Analyze + document
=====
type: prompt
Complete the following tasks and **write instructions / documentation** for your
collaborator to reproduce your work starting with the original dataset
(`data/gapminder-5060.csv`).
1. Download material: http://bit.ly/cbb-retreat -> [Releases](https://github.com/Reproducible-Science-Curriculum/cbb-retreat/releases/) -> Latest
2. Visualize life expectancy over time for Canada in the 1950s and 1960s using
a line plot.
3. Something is clearly wrong with this plot! Turns out there's a data error
in the data file: life expectancy for Canada in the year 1957 is coded
as `999999`, it should actually be `69.96`. Make this correction.
4. Visualize life expectancy over time for Canada again, with the corrected data.<br>
*Stretch goal:* Add lines for Mexico and United States.
Part 2: Swap + discuss
=====
type: prompt
Introduce yourself to your collaborator.
1. Swap instructions / documentation with your collaborator, and try to reproduce
their work, first **without talking to each oher**.
If your collaborator does not have the software they need to reproduce your work, we
encourage you to either help them install it or walk them through it on your computer
in a way that would emulate the experience. (Remember, this could be part of the
irreproducibility problem!)
2. Then, talk to each other about challenges you faced (or didn't face) or why
you were or weren't able to reproduce their work.
Reflection
=====
type: prompt
This exercise:
- What tools did you use (Excel, R / Python, Word / plain text etc.)?
- What made it easy / hard for reproducing your partners' work?
In a lab setting:
- What would happen if your collaborator is no longer available to walk you through
their analysis?
- What would have to happen if you
- had to swap out the dataset or extend the analysis?
- caught further errors and had to re-create the analysis?
- you had to revert back to the original dataset?
Four facets of reproducibility
=====
type: titleonly
Four facets of reproducibility
=====
1. **Documentation:** difference between binary files (e.g. docx) and text files and
why text files are preferred for documentation
- *Protip:* Use markdown to document your workflow so that anyone can pick up your
data and follow what you are doing
- *Protip:* Use literate programming so that your analysis and your results are
tightly connected, or better yet, unseperable
2. **Organization:** tools to organize your projects so that you don't have a single
folder with hundreds of files
3. **Automation:** the power of scripting to create automated data analyses
4. **Dissemination:** publishing is not the end of your analysis, rather it is a way
station towards your future research and the future research of others
Executable documentation
=====
type: titleonly
Literate Programming
=====
Provenance with results pasted into manuscript:

* Which code?
* Which data?
* Which context?
***
vs. Provenance of figures with Rmarkdown reports:
[](https://github.com/Reproducible-Science-Curriculum/rr-organization1/raw/master/files/lit-prog/countryPick4.pdf)
Demo - Literate Programming
=====
type: titleonly
Executable data integrity checks
=====
<br/>
```{r, echo=FALSE}
gap_5060 <- read.csv("material/data/gapminder-5060.csv")
```
Life expectancy shouldn't exceed even the most extreme age observed for humans.
```{r, error=TRUE}
if (any(gap_5060$lifeExp > 150)) {
stop("improbably high life expectancies")
}
```
Executable data integrity checks
=====
<br/>
The library `testthat` allows us to make this a little more readable:
```{r, error=TRUE}
library(testthat)
expect_that(any(gap_5060$lifeExp > 150), is_false(),
"improbably high life expectancies")
```
Naming your files
=====
type: titleonly
Research projects grow
=====
<br/><br/>
- There are going to be files. _Lots of files._
- They will change over time.
- They will have differing relationships to each other.
File organization and naming are effective weapons against chaos.
Three key principles for (file) names
=====
* Machine readable
- easy to search for files later
- easy to narrow file lists based on names
- easy to extract info from file names (regexp-friendly)
* Human readable
- name contains information on content, or
- name contains semantics (e.g., place in workflow)
* Plays well with default ordering
- use numeric prefix to induce logic order
- left pad numbers with zeros
- use ISO 8601 standard (YYYY-mm-dd) for dates
Exercise
=====
type: prompt
<br/><br/>
Your data files contain readings from a well plate, one file per well,
using a specific assay run on a certain date, after a certain treatment.
* Devise a naming scheme for the files that is both "machine" and "human" readable.
Example:
=====
```sh
$ ls *Plsmd*
2013-06-26_BRAFASSAY_Plsmd-CL56-1MutFrac_A01.csv
2013-06-26_BRAFASSAY_Plsmd-CL56-1MutFrac_A02.csv
2013-06-26_BRAFASSAY_Plsmd-CL56-1MutFrac_A03.csv
2013-06-26_BRAFASSAY_Plsmd-CL56-1MutFrac_B01.csv
2013-06-26_BRAFASSAY_Plsmd-CL56-1MutFrac_B02.csv
...
2013-06-26_BRAFASSAY_Plsmd-CL56-1MutFrac_H03.csv
```
```{r, eval=FALSE}
> list.files(pattern = "Plsmd") %>% head
[1] 2013-06-26_BRAFASSAY_Plsmd-CL56-1MutFrac_A01.csv
[2] 2013-06-26_BRAFASSAY_Plsmd-CL56-1MutFrac_A02.csv
[3] 2013-06-26_BRAFASSAY_Plsmd-CL56-1MutFrac_A03.csv
[4] 2013-06-26_BRAFASSAY_Plsmd-CL56-1MutFrac_B01.csv
[5] 2013-06-26_BRAFASSAY_Plsmd-CL56-1MutFrac_B02.csv
[6] 2013-06-26_BRAFASSAY_Plsmd-CL56-1MutFrac_B03.csv
```
Example:
=====
```{r, echo=FALSE, results="hide"}
flist <- c("2013-06-26_BRAFASSAY_Plsmd-CL56-1MutFrac_A01.csv",
"2013-06-26_BRAFASSAY_Plsmd-CL56-1MutFrac_A02.csv",
"2013-06-26_BRAFASSAY_Plsmd-CL56-1MutFrac_A03.csv",
"2013-06-26_BRAFASSAY_Plsmd-CL56-1MutFrac_B01.csv",
"2013-06-26_BRAFASSAY_Plsmd-CL56-1MutFrac_B02.csv",
"2013-06-26_BRAFASSAY_Plsmd-CL56-1MutFrac_B03.csv")
```
```{r}
meta <- stringr::str_split_fixed(flist, "[_\\.]", 5)
colnames(meta) <-
c("date", "assay", "experiment", "well", "ext")
meta[,1:4]
```
Organizing your files
=====
type: titleonly
Best practices for file organization
=====
class: centered-image
[](https://doi.org/10.1371/journal.pcbi.1000424)
<small>Noble, William Stafford. 2009. “[A Quick Guide to Organizing Computational Biology Projects](https://doi.org/10.1371/journal.pcbi.1000424).” PLoS Computational Biology 5 (7): e1000424.</small>
A data science project using R
=====
title: false
```
|
+-- data-raw/
| |
| +-- gapminder-5060.csv
| +-- gapminder-7080.csv.csv
| +-- ....
|
+-- data-output/
|
+-- fig/
|
+-- R/
| |
| +-- figures.R
| +-- data.R
| +-- utils.R
| +-- dependencies.R
|
+-- tests/
|
+-- manuscript.Rmd
+-- make.R
```
Some principles for file organization
=====
- `data-raw`: the original data, you shouldn't edit or otherwise alter any of
the files in this folder.
- `data-output`: intermediate datasets that will be generated by the
analysis.
- We write them to CSV files so we could share or archive them,
for example if they take a long time (or expensive resources) to generate.
- `fig`: the folder where we can store the figures used in the manuscript.
- `R`: our R code (the functions)
- Often easier to keep the prose separated from the code.
- If you have a lot of code (and/or manuscript is long), it's easier
to navigate.
- `tests`: the code to test that our functions are behaving properly and that
all our data is included in the analysis.
Automation
=====
type: titleonly
Make everything
=====
```{r}
make_ms <- function() {
rmarkdown::render("manuscript.Rmd",
"html_document")
invisible(file.exists("manuscript.html"))
}
clean_ms <- function() {
res <- file.remove("manuscript.html")
invisible(res)
}
make_all <- function() {
make_data()
make_figures()
make_tests()
make_ms()
}
clean_all <- function() {
clean_data()
clean_figures()
clean_ms()
}
```
Tests made easier
=====
`testthat` includes a function called `test_dir` that will run tests
included in files in a given directory. We can use it to run all the tests in
our `tests/` folder.
```{r, eval=FALSE}
test_dir("tests/")
```
Let's turn it into a function, so we'll be able to add some additional
functionalities to it a little later. We are also going to save it at the root
of our working directory in the file called `make.R`:
```{r}
## add this to make.R
make_tests <- function() {
test_dir("tests/")
}
```
Dissemination - Sharing, publishing, archiving
=====
type: titleonly
Why share / archive data & code?
=====
incremental: true
* funding agency / journal requirement
* community expects it
* _increased visibility / citation_
increased visibility / citation
=====
left: 70%
[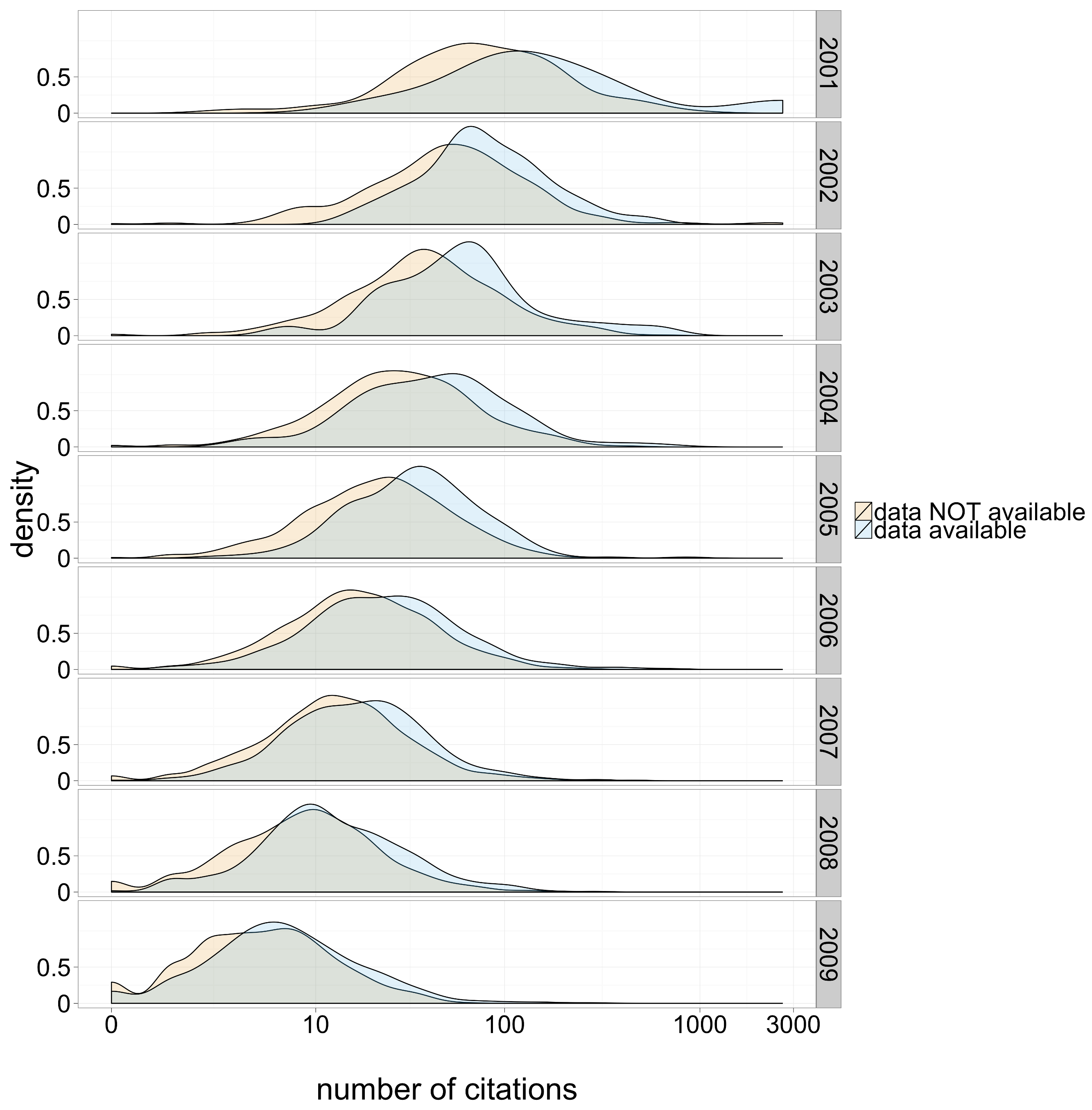](https://doi.org/10.7717/peerj.175)
***
Piwowar & Vision (2013) "[Data reuse and the open data citation advantage.](https://doi.org/10.7717/peerj.175)" PeerJ, e175
<small>Figure 1: Citation density for papers with and without publicly available microarray data, by year of study publication.</small>
Why share / archive data & code?
=====
* funding agency / journal requirement
* community expects it
* increased visibility / citation
* _better research_
Better research
=====
left: 70%
[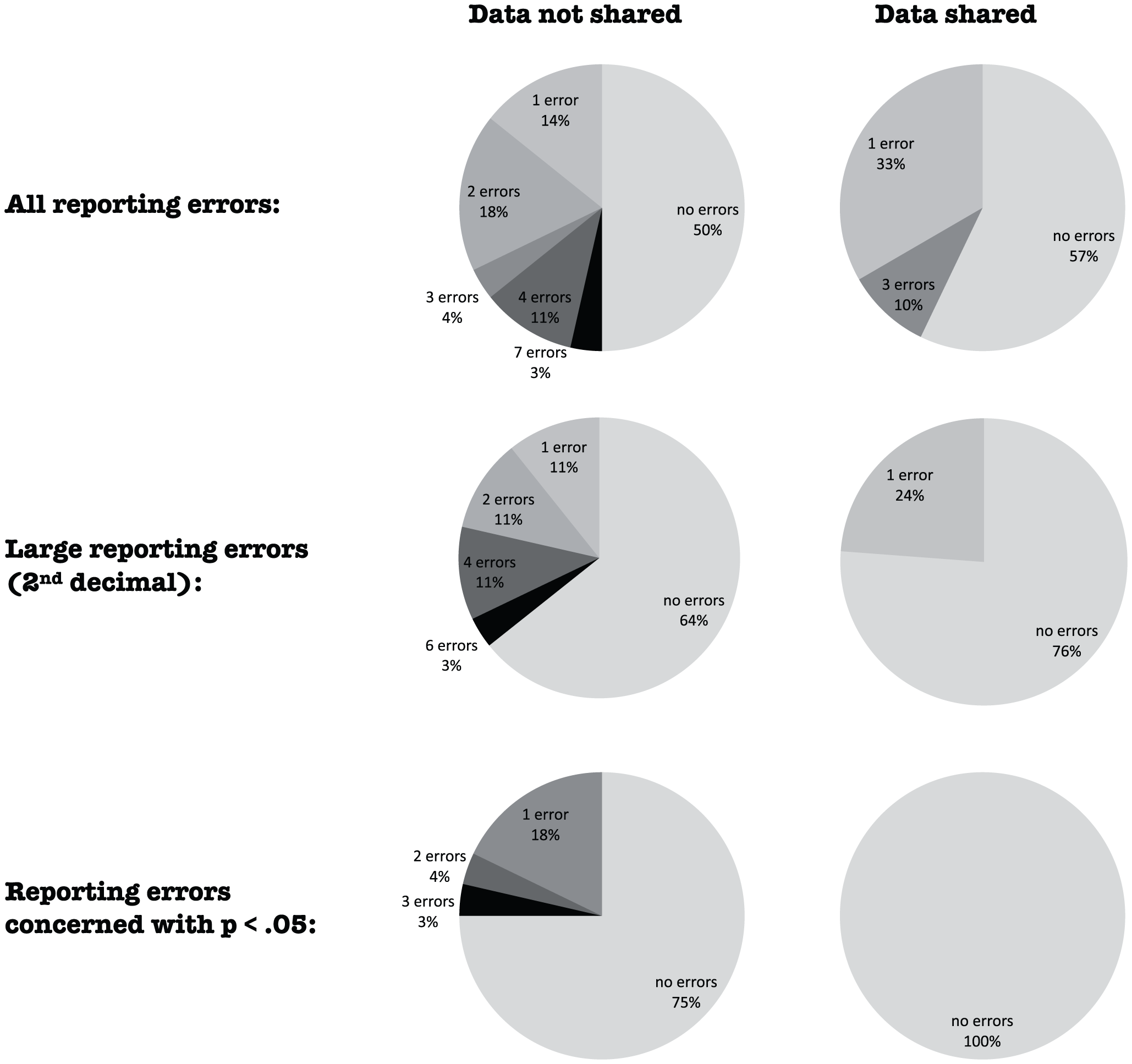](https://doi.org/10.1371/journal.pone.0026828)
***
<small>Wicherts et al (2011) "[Willingness to Share Research Data Is Related to the Strength of the Evidence and the Quality of Reporting of Statistical Results.](https://doi.org/10.1371/journal.pone.0026828)" PLoS ONE 6(11): e26828</small>
<p style="font-size: 75%">Figure 1. Distribution of reporting errors per paper for papers from which data were shared and from which no data were shared.</p>
Where archive & publish?
=====
* Domain-specific data repository (GenBank, PDB)
* Source code hosting service ([GitHub](http://github.com), [Bitbucket](http://bitbucket.com))
* Generic repository ([Dryad](http://datadryad.com), [Figshare](http://figshare.com), [Zenodo](http://zenodo.org))
* Institutional repository
* Journal supplementary materials
Where archive & publish?
=====
* Domain-specific data repository (GenBank, PDB)
* Source code hosting service ([GitHub](http://github.com), [Bitbucket](http://bitbucket.com))
* Generic repository ([Dryad](http://datadryad.com), [Figshare](http://figshare.com), [Zenodo](http://zenodo.org))
* Institutional repository
* ~~Journal supplementary materials~~
How to choose a repository
=====
* Is there a domain specific repository?
* What are the backup & replication policies?
* Is there a plan for long-term preservation?
* Can people find your materials?
* Is it citable? (does it provide DOIs)
University libraries try to help
=====
title: false
<!-- Replace with your university library's data management guide -->
<iframe width="100%" height="700px" src="http://library.duke.edu/data/guides/data-management"></iframe>
How to share, publish: file formats
=====
**Do's**
* non-proprietary file formats
* text file formats (.csv, .tsv, .txt)
***
**Don't's**
* proprietary file formats (.xls)
* data as PDFs or images
* data in Word documents
Put a license on creative works
=====
type: titleonly
Software licensing guide
=====
left: 70%
[](10.1371/journal.pcbi.1002598.g002)
***
> Morin, Andrew, Jennifer Urban, and Piotr Sliz. 2012. “[A Quick Guide to Software Licensing for the Scientist-Programmer.](https://doi.org/10.1371/journal.pcbi.1002598)” PLoS Computational Biology 8 (7): e1002598.
Don't put a license on facts (a.k.a. data)
=====
type: titleonly
Licenses versus community norms
=====
<center>[](http://creativecommons.org/about/cc0)</center>
From the [Panton Principles](http://pantonprinciples.org/faq/#Q11_What_are_community_norms_and_why_are_they_important):
> [In] the scholarly research community the act of citation is a commonly held community norm when reusing another community member’s work. [...] A well functioning community supports its members in their application of norms, whereas licences can only be enforced through court action and thus invite people to ignore them when they are confident that this is unlikely.
Good - Better - Best
=====
<br/>
<br/>
[](https://doi.org/10.1126/science.1213847)
Peng, R. D. “[Reproducible Research in Computational Science](https://doi.org/10.1126/science.1213847)” Science 334, no. 6060 (2011): 1226–1227
Forming reproducible habits pays off
=====
incremental: true
* Reproducible practices can be applied after the fact, but it's much harder.
- And now you're doing this for others, rather than for your own benefits.
- And seriously, you won't ever publish the stuff you're working on?
* Adopting practices for reproducible science from the outset pays off in multiple ways.
- It's easy and little work while the project is still small and contains few files.
- Now all you're doing is reproducible. No painful considerations when it comes to sharing stuff.
- Your future self will reap the benefits.
- (Your lab does, too, but that's a side effect.)
- (Who should be leading this - you or your old-school advisor?)
Acknowledgements
=====
* Organizing committee members and _curriculum instructors_
* _Mine Çetinkaya-Rundel_ (Duke)
* _Karen Cranston_ (Duke)
* _Ciera Martinez_ (UC Davis)
* _François Michonneau_ (iDigBio, U. Florida)
* Matt Pennell (U. Idaho)
* Tracy Teal (Data Carpentry)
* _Dan Leehr_ (Duke)
***
* Inspiration: Sophie (Kershaw) Kay - [Rotation Based Learning](http://www.opensciencetraining.com/content.php)
* Funding and support:
* US National Science Foundation (NSF)
* National Evolutionary Synthesis Center (NESCent)
* Center for Genomic & Computational Biology (GCB), Duke University
Eating our dogfood: text formats, version control, sharing
=====
<p/>
This slideshow was generated as HTML from Markdown using RStudio.
The Markdown sources, and the HTML, are hosted on Github:
https://github.com/Reproducible-Science-Curriculum/cbb-retreat
***
<p/>

[](https://www.rstudio.com)
Further Resources
=====
* Entire [suppl. doc](http://www.sciencemag.org/content/suppl/2015/04/29/348.6234.567.DC1/Finnegan.SM.pdf) generated from Rmarkdown: Finnegan et al. 2015. “[Paleontological Baselines for Evaluating Extinction Risk in the Modern Oceans](https://doi.org/10.1126/science.aaa6635).” Science 348 (6234): 567–70.
* [Data Analysis for the Life Sciences](http://simplystatistics.org/?p=4311) - a book completely written in R markdown
* FitzJohn et al. 2014. “[How Much of the World Is Woody?](https://doi.org/10.1111/1365-2745.12260)” The Journal of Ecology. doi:10.1111/1365-2745.12260. [Start to end replicable analysis on Github](https://github.com/richfitz/wood).
* Boettiger et al. “[RNeXML: A Package for Reading and Writing Richly Annotated Phylogenetic, Character, and Trait Data in R](https://doi.org/10.1111/2041-210X.12469).” Methods in Ecology and Evolution, September. [Code archive and DOI assignment at Zenodo](https://doi.org/10.5281/zenodo.13131)