+
Albadry, Mohamed, Sebastian Höpfl, Nadia Ehteshamzad, Matthias König, Michael Böttcher, Jasna Neumann, Amelie Lupp, et al. 2022. “Periportal Steatosis in Mice Affects Distinct Parameters of Pericentral Drug Metabolism.” Scientific Reports 12 (1): 21825. https://doi.org/10.1038/s41598-022-26483-6.
+
Fröhlich, Fabian, Luca Gerosa, Jeremy Muhlich, and Peter K. Sorger. 2022. “Mechanistic Model of Mapk Signaling Reveals How Allostery and Rewiring Contribute to Drug Resistance.” bioRxiv. https://doi.org/10.1101/2022.02.17.480899.
-
Lakrisenko, Polina, Paul Stapor, Stephan Grein, Łukasz Paszkowski, Dilan Pathirana, Fabian Fröhlich, Glenn Terje Lines, Daniel Weindl, and Jan Hasenauer. 2022. “Efficient Computation of Adjoint Sensitivities at Steady-State in ODE Models of Biochemical Reaction Networks.” bioRxiv. https://doi.org/10.1101/2022.08.08.503176.
+
+
+
Meyer, Kristian, Mikkel Søes Ibsen, Lisa Vetter-Joss, Ernst Broberg Hansen, and Jens Abildskov. 2022. “Industrial Ion-Exchange Chromatography Development Using Discontinuous Galerkin Methods Coupled with Forward Sensitivity Analysis.” Journal of Chromatography A, 463741. https://doi.org/10.1016/j.chroma.2022.463741.
+
+
Schmucker, Robin, Gabriele Farina, James Faeder, Fabian Fröhlich, Ali Sinan Saglam, and Tuomas Sandholm. 2022. “Combination Treatment Optimization Using a Pan-Cancer Pathway Model.” PLOS Computational Biology 17 (12): 1–22. https://doi.org/10.1371/journal.pcbi.1009689.
@@ -27,8 +39,8 @@ If you applied AMICI in your work and your publication is missing, please let us
Stapor, Paul, Leonard Schmiester, Christoph Wierling, Simon Merkt, Dilan Pathirana, Bodo M. H. Lange, Daniel Weindl, and Jan Hasenauer. 2022. “Mini-batch optimization enables training of ODE models on large-scale datasets.” Nature Communications 13 (1): 34. https://doi.org/10.1038/s41467-021-27374-6.
-
-
Sundqvist, Nicolas, Sebastian Sten, Maria Engström, and Gunnar Cedersund. 2022. “Mechanistic Model for Human Brain Metabolism and the Neurovascular Coupling.” bioRxiv. https://doi.org/10.1101/2022.02.15.480629.
+
+
Sundqvist, Nicolas, Sebastian Sten, Peter Thompson, Benjamin Jan Andersson, Maria Engström, and Gunnar Cedersund. 2022. “Mechanistic Model for Human Brain Metabolism and Its Connection to the Neurovascular Coupling.” PLOS Computational Biology 18 (12): 1–24. https://doi.org/10.1371/journal.pcbi.1010798.
Villaverde, Alejandro F., Elba Raimúndez, Jan Hasenauer, and Julio R. Banga. 2022. “Assessment of Prediction Uncertainty Quantification Methods in Systems Biology.” IEEE/ACM Transactions on Computational Biology and Bioinformatics, 1–12. https://doi.org/10.1109/TCBB.2022.3213914.
diff --git a/documentation/rtd_requirements.txt b/documentation/rtd_requirements.txt
index 37d6f7663c..c5fe2d49a9 100644
--- a/documentation/rtd_requirements.txt
+++ b/documentation/rtd_requirements.txt
@@ -1,3 +1,4 @@
+# NOTE: relative paths are expected to be relative to the repository root
sphinx==5.1.1
mock>=4.0.3
setuptools==65.5.1
@@ -20,3 +21,6 @@ sphinxcontrib-napoleon>=0.7
pygments==2.13.0
Jinja2==3.1.2
git+https://github.com/readthedocs/readthedocs-sphinx-ext
+ipykernel
+-e git+https://github.com/Benchmarking-Initiative/Benchmark-Models-PEtab.git@master#subdirectory=src/python&egg=benchmark_models_petab
+-e python/sdist/
diff --git a/include/amici/amici.h b/include/amici/amici.h
index 3f022361c3..1201b3b1b7 100644
--- a/include/amici/amici.h
+++ b/include/amici/amici.h
@@ -20,10 +20,9 @@ namespace amici {
* @param rethrow rethrow integration exceptions?
* @return rdata pointer to return data object
*/
-std::unique_ptr
runAmiciSimulation(Solver &solver,
- const ExpData *edata,
- Model &model,
- bool rethrow = false);
+std::unique_ptr runAmiciSimulation(
+ Solver& solver, ExpData const* edata, Model& model, bool rethrow = false
+);
/**
* @brief Same as runAmiciSimulation, but for multiple ExpData instances. When
@@ -36,15 +35,15 @@ std::unique_ptr runAmiciSimulation(Solver &solver,
* @param num_threads number of threads for parallel execution
* @return vector of pointers to return data objects
*/
-std::vector>
-runAmiciSimulations(Solver const &solver, const std::vector &edatas,
- Model const &model, bool failfast, int num_threads);
-
+std::vector> runAmiciSimulations(
+ Solver const& solver, std::vector const& edatas,
+ Model const& model, bool failfast, int num_threads
+);
/**
* @brief Get the string representation of the given simulation status code
* (see ReturnData::status).
- * @param return_code
+ * @param status Status code
* @return Name of the variable representing this status code.
*/
std::string simulation_status_to_str(int status);
diff --git a/include/amici/defines.h b/include/amici/defines.h
index e8321e3714..02437f77cb 100644
--- a/include/amici/defines.h
+++ b/include/amici/defines.h
@@ -71,6 +71,7 @@ constexpr int AMICI_TOO_MUCH_ACC= -2;
constexpr int AMICI_ERR_FAILURE= -3;
constexpr int AMICI_CONV_FAILURE= -4;
constexpr int AMICI_RHSFUNC_FAIL= -8;
+constexpr int AMICI_FIRST_RHSFUNC_ERR= -9;
constexpr int AMICI_ILL_INPUT= -22;
constexpr int AMICI_ERROR= -99;
constexpr int AMICI_NO_STEADY_STATE= -81;
diff --git a/include/amici/edata.h b/include/amici/edata.h
index b15e34fc10..1f18cea991 100644
--- a/include/amici/edata.h
+++ b/include/amici/edata.h
@@ -28,7 +28,7 @@ class ExpData : public SimulationParameters {
* @brief Copy constructor, needs to be declared to be generated in
* swig
*/
- ExpData(const ExpData &) = default;
+ ExpData(ExpData const&) = default;
/**
* @brief constructor that only initializes dimensions
@@ -88,7 +88,7 @@ class ExpData : public SimulationParameters {
*
* @param model pointer to model specification object
*/
- explicit ExpData(const Model &model);
+ explicit ExpData(Model const& model);
/**
* @brief constructor that initializes with returnData, adds noise according
@@ -98,7 +98,7 @@ class ExpData : public SimulationParameters {
* @param sigma_y scalar standard deviations for all observables
* @param sigma_z scalar standard deviations for all event observables
*/
- ExpData(const ReturnData &rdata, realtype sigma_y, realtype sigma_z);
+ ExpData(ReturnData const& rdata, realtype sigma_y, realtype sigma_z);
/**
* @brief constructor that initializes with returnData, adds noise according
@@ -110,12 +110,14 @@ class ExpData : public SimulationParameters {
* @param sigma_z vector of standard deviations for event observables
* (dimension: nztrue or nmaxevent x nztrue, row-major)
*/
- ExpData(const ReturnData &rdata, std::vector sigma_y,
- std::vector sigma_z);
+ ExpData(
+ ReturnData const& rdata, std::vector sigma_y,
+ std::vector sigma_z
+ );
~ExpData() = default;
- friend inline bool operator==(const ExpData& lhs, const ExpData& rhs);
+ friend inline bool operator==(ExpData const& lhs, ExpData const& rhs);
/**
* @brief number of observables of the non-augmented model
@@ -150,7 +152,7 @@ class ExpData : public SimulationParameters {
*
* @param ts timepoints
*/
- void setTimepoints(const std::vector &ts);
+ void setTimepoints(std::vector const& ts);
/**
* @brief get function that copies data from ExpData::ts to output
@@ -173,7 +175,7 @@ class ExpData : public SimulationParameters {
*
* @param observedData observed data (dimension: nt x nytrue, row-major)
*/
- void setObservedData(const std::vector &observedData);
+ void setObservedData(std::vector const& observedData);
/**
* @brief set function that copies observed data for specific observable
@@ -181,7 +183,7 @@ class ExpData : public SimulationParameters {
* @param observedData observed data (dimension: nt)
* @param iy observed data index
*/
- void setObservedData(const std::vector &observedData, int iy);
+ void setObservedData(std::vector const& observedData, int iy);
/**
* @brief get function that checks whether data at specified indices has
@@ -208,7 +210,7 @@ class ExpData : public SimulationParameters {
*
* @return pointer to observed data at index (dimension: nytrue)
*/
- const realtype *getObservedDataPtr(int it) const;
+ realtype const* getObservedDataPtr(int it) const;
/**
* @brief set function that copies data from input to
@@ -217,7 +219,7 @@ class ExpData : public SimulationParameters {
* @param observedDataStdDev standard deviation of observed data (dimension:
* nt x nytrue, row-major)
*/
- void setObservedDataStdDev(const std::vector &observedDataStdDev);
+ void setObservedDataStdDev(std::vector const& observedDataStdDev);
/**
* @brief set function that sets all ExpData::observedDataStdDev to the
@@ -235,8 +237,9 @@ class ExpData : public SimulationParameters {
* nt)
* @param iy observed data index
*/
- void setObservedDataStdDev(const std::vector &observedDataStdDev,
- int iy);
+ void setObservedDataStdDev(
+ std::vector const& observedDataStdDev, int iy
+ );
/**
* @brief set function that sets all standard deviation of a specific
@@ -272,7 +275,7 @@ class ExpData : public SimulationParameters {
* @param it timepoint index
* @return pointer to standard deviation of observed data at index
*/
- const realtype *getObservedDataStdDevPtr(int it) const;
+ realtype const* getObservedDataStdDevPtr(int it) const;
/**
* @brief set function that copies observed event data from input to
@@ -281,7 +284,7 @@ class ExpData : public SimulationParameters {
* @param observedEvents observed data (dimension: nmaxevent x nztrue,
* row-major)
*/
- void setObservedEvents(const std::vector &observedEvents);
+ void setObservedEvents(std::vector const& observedEvents);
/**
* @brief set function that copies observed event data for specific event
@@ -290,7 +293,7 @@ class ExpData : public SimulationParameters {
* @param observedEvents observed data (dimension: nmaxevent)
* @param iz observed event data index
*/
- void setObservedEvents(const std::vector &observedEvents, int iz);
+ void setObservedEvents(std::vector const& observedEvents, int iz);
/**
* @brief get function that checks whether event data at specified indices
@@ -317,7 +320,7 @@ class ExpData : public SimulationParameters {
*
* @return pointer to observed event data at ieth occurrence
*/
- const realtype *getObservedEventsPtr(int ie) const;
+ realtype const* getObservedEventsPtr(int ie) const;
/**
* @brief set function that copies data from input to
@@ -326,7 +329,7 @@ class ExpData : public SimulationParameters {
* @param observedEventsStdDev standard deviation of observed event data
*/
void
- setObservedEventsStdDev(const std::vector &observedEventsStdDev);
+ setObservedEventsStdDev(std::vector const& observedEventsStdDev);
/**
* @brief set function that sets all ExpData::observedDataStdDev to the
@@ -344,9 +347,9 @@ class ExpData : public SimulationParameters {
* (dimension: nmaxevent)
* @param iz observed data index
*/
- void
- setObservedEventsStdDev(const std::vector &observedEventsStdDev,
- int iz);
+ void setObservedEventsStdDev(
+ std::vector const& observedEventsStdDev, int iz
+ );
/**
* @brief set function that sets all standard deviation of a specific
@@ -384,7 +387,7 @@ class ExpData : public SimulationParameters {
* @return pointer to standard deviation of observed event data at ie-th
* occurrence
*/
- const realtype *getObservedEventsStdDevPtr(int ie) const;
+ realtype const* getObservedEventsStdDevPtr(int ie) const;
/**
* @brief Arbitrary (not necessarily unique) identifier.
@@ -414,8 +417,9 @@ class ExpData : public SimulationParameters {
* @param input vector input to be checked
* @param fieldname name of the input
*/
- void checkDataDimension(std::vector const &input,
- const char *fieldname) const;
+ void checkDataDimension(
+ std::vector const& input, char const* fieldname
+ ) const;
/**
* @brief checker for dimensions of input observedEvents or
@@ -424,8 +428,9 @@ class ExpData : public SimulationParameters {
* @param input vector input to be checked
* @param fieldname name of the input
*/
- void checkEventsDimension(std::vector const &input,
- const char *fieldname) const;
+ void checkEventsDimension(
+ std::vector const& input, char const* fieldname
+ ) const;
/** @brief number of observables */
int nytrue_{0};
@@ -457,33 +462,34 @@ class ExpData : public SimulationParameters {
std::vector observed_events_std_dev_;
};
-
-inline bool operator==(const ExpData& lhs, const ExpData& rhs) {
- return *dynamic_cast< const SimulationParameters* >(&lhs)
- == *dynamic_cast< const SimulationParameters* >(&rhs)
- && lhs.id == rhs.id
- && lhs.nytrue_ == rhs.nytrue_
- && lhs.nztrue_ == rhs.nztrue_
- && lhs.nmaxevent_ == rhs.nmaxevent_
- && is_equal(lhs.observed_data_,
- rhs.observed_data_)
- && is_equal(lhs.observed_data_std_dev_,
- rhs.observed_data_std_dev_)
- && is_equal(lhs.observed_events_,
- rhs.observed_events_)
- && is_equal(lhs.observed_events_std_dev_,
- rhs.observed_events_std_dev_);
+/**
+ * @brief Equality operator
+ * @param lhs some object
+ * @param rhs another object
+ * @return `true`, if both arguments are equal; `false` otherwise.
+ */
+inline bool operator==(ExpData const& lhs, ExpData const& rhs) {
+ return *dynamic_cast(&lhs)
+ == *dynamic_cast(&rhs)
+ && lhs.id == rhs.id && lhs.nytrue_ == rhs.nytrue_
+ && lhs.nztrue_ == rhs.nztrue_ && lhs.nmaxevent_ == rhs.nmaxevent_
+ && is_equal(lhs.observed_data_, rhs.observed_data_)
+ && is_equal(lhs.observed_data_std_dev_, rhs.observed_data_std_dev_)
+ && is_equal(lhs.observed_events_, rhs.observed_events_)
+ && is_equal(
+ lhs.observed_events_std_dev_, rhs.observed_events_std_dev_
+ );
};
-
/**
* @brief checks input vector of sigmas for not strictly positive values
*
* @param sigmaVector vector input to be checked
* @param vectorName name of the input
*/
-void checkSigmaPositivity(std::vector const &sigmaVector,
- const char *vectorName);
+void checkSigmaPositivity(
+ std::vector const& sigmaVector, char const* vectorName
+);
/**
* @brief checks input scalar sigma for not strictly positive value
@@ -491,7 +497,7 @@ void checkSigmaPositivity(std::vector const &sigmaVector,
* @param sigma input to be checked
* @param sigmaName name of the input
*/
-void checkSigmaPositivity(realtype sigma, const char *sigmaName);
+void checkSigmaPositivity(realtype sigma, char const* sigmaName);
/**
* @brief The ConditionContext class applies condition-specific amici::Model
@@ -508,10 +514,11 @@ class ConditionContext : public ContextManager {
* @param fpc flag indicating which fixedParameter from edata to apply
*/
explicit ConditionContext(
- Model *model, const ExpData *edata = nullptr,
- FixedParameterContext fpc = FixedParameterContext::simulation);
+ Model* model, ExpData const* edata = nullptr,
+ FixedParameterContext fpc = FixedParameterContext::simulation
+ );
- ConditionContext &operator=(const ConditionContext &other) = delete;
+ ConditionContext& operator=(ConditionContext const& other) = delete;
~ConditionContext();
@@ -523,8 +530,7 @@ class ConditionContext : public ContextManager {
* @param edata
* @param fpc flag indicating which fixedParameter from edata to apply
*/
- void applyCondition(const ExpData *edata,
- FixedParameterContext fpc);
+ void applyCondition(ExpData const* edata, FixedParameterContext fpc);
/**
* @brief Restore original settings on constructor-supplied amici::Model.
diff --git a/include/amici/exception.h b/include/amici/exception.h
index 5f35324d41..7ee91d511b 100644
--- a/include/amici/exception.h
+++ b/include/amici/exception.h
@@ -18,8 +18,9 @@ class AmiException : public std::exception {
public:
/**
* @brief Default ctor.
+ * @param first_frame Index of first frame to include
*/
- AmiException();
+ AmiException(int const first_frame = 3);
/**
* @brief Constructor with printf style interface
@@ -43,8 +44,9 @@ class AmiException : public std::exception {
/**
* @brief Stores the current backtrace
* @param nMaxFrames number of frames to go back in stacktrace
+ * @param first_frame Index of first frame to include
*/
- void storeBacktrace(int nMaxFrames);
+ void storeBacktrace(int nMaxFrames, int const first_frame);
protected:
/**
diff --git a/include/amici/forwardproblem.h b/include/amici/forwardproblem.h
index d1aa49caa7..581361dc6c 100644
--- a/include/amici/forwardproblem.h
+++ b/include/amici/forwardproblem.h
@@ -33,8 +33,10 @@ class ForwardProblem {
* @param preeq preequilibration with which to initialize the forward problem,
* pass nullptr for no initialization
*/
- ForwardProblem(const ExpData *edata, Model *model, Solver *solver,
- const SteadystateProblem *preeq);
+ ForwardProblem(
+ ExpData const* edata, Model* model, Solver* solver,
+ SteadystateProblem const* preeq
+ );
~ForwardProblem() = default;
@@ -53,7 +55,7 @@ class ForwardProblem {
* @param model Model instance
* @param edata experimental data
*/
- void getAdjointUpdates(Model &model, const ExpData &edata);
+ void getAdjointUpdates(Model& model, ExpData const& edata);
/**
* @brief Accessor for t
@@ -221,7 +223,7 @@ class ForwardProblem {
* @param it timepoint index
* @return state
*/
- const SimulationState &getSimulationStateTimepoint(int it) const {
+ SimulationState const& getSimulationStateTimepoint(int it) const {
if (model->getTimepoint(it) == initial_state_.t)
return getInitialSimulationState();
return timepoint_states_.find(model->getTimepoint(it))->second;
@@ -233,7 +235,7 @@ class ForwardProblem {
* @param iroot event index
* @return SimulationState
*/
- const SimulationState &getSimulationStateEvent(int iroot) const {
+ SimulationState const& getSimulationStateEvent(int iroot) const {
return event_states_.at(iroot);
};
@@ -242,7 +244,7 @@ class ForwardProblem {
* initial timepoint
* @return SimulationState
*/
- const SimulationState &getInitialSimulationState() const {
+ SimulationState const& getInitialSimulationState() const {
return initial_state_;
};
@@ -251,7 +253,7 @@ class ForwardProblem {
* final timepoint (or when simulation failed)
* @return SimulationState
*/
- const SimulationState &getFinalSimulationState() const {
+ SimulationState const& getFinalSimulationState() const {
return final_state_;
};
@@ -262,7 +264,7 @@ class ForwardProblem {
Solver *solver;
/** pointer to experimental data instance */
- const ExpData *edata;
+ ExpData const* edata;
private:
@@ -293,8 +295,6 @@ class ForwardProblem {
/**
* @brief Applies the event bolus to the current state
- *
- * @param model pointer to model specification object
*/
void applyEventBolus();
@@ -451,7 +451,7 @@ class FinalStateStorer : public ContextManager {
explicit FinalStateStorer(ForwardProblem *fwd) : fwd_(fwd) {
}
- FinalStateStorer &operator=(const FinalStateStorer &other) = delete;
+ FinalStateStorer& operator=(FinalStateStorer const& other) = delete;
/**
* @brief destructor, stores simulation state
diff --git a/include/amici/misc.h b/include/amici/misc.h
index fe9fb5b8e2..91c26cffa0 100644
--- a/include/amici/misc.h
+++ b/include/amici/misc.h
@@ -45,8 +45,7 @@ gsl::span slice(std::vector &data, int index, unsigned size) {
*/
template
-gsl::span slice(const std::vector &data,
- int index, unsigned size) {
+gsl::span slice(std::vector const& data, int index, unsigned size) {
if ((index + 1) * size > data.size())
throw std::out_of_range("requested slice is out of data range");
if (size > 0)
@@ -78,7 +77,7 @@ void checkBufferSize(gsl::span buffer,
* @param buffer buffer to which values are to be written
*/
template
-void writeSlice(const gsl::span slice, gsl::span buffer) {
+void writeSlice(const gsl::span slice, gsl::span buffer) {
checkBufferSize(buffer, slice.size());
std::copy(slice.begin(), slice.end(), buffer.data());
};
@@ -89,7 +88,7 @@ void writeSlice(const gsl::span slice, gsl::span buffer) {
* @param buffer buffer to which values are to be added
*/
template
-void addSlice(const gsl::span slice, gsl::span buffer) {
+void addSlice(const gsl::span slice, gsl::span buffer) {
checkBufferSize(buffer, slice.size());
std::transform(slice.begin(), slice.end(), buffer.begin(), buffer.begin(),
std::plus());
@@ -100,8 +99,7 @@ void addSlice(const gsl::span slice, gsl::span buffer) {
* @param s computed value
* @param b buffer to which values are to be written
*/
-template
-void writeSlice(const std::vector &s, std::vector &b) {
+template void writeSlice(std::vector const& s, std::vector& b) {
writeSlice(gsl::make_span(s.data(), s.size()),
gsl::make_span(b.data(), b.size()));
};
@@ -111,8 +109,7 @@ void writeSlice(const std::vector &s, std::vector &b) {
* @param s computed value
* @param b buffer to which values are to be written
*/
-template
-void writeSlice(const std::vector &s, gsl::span b) {
+template void writeSlice(std::vector const& s, gsl::span b) {
writeSlice(gsl::make_span(s.data(), s.size()), b);
};
@@ -121,8 +118,7 @@ void writeSlice(const std::vector &s, gsl::span b) {
* @param s computed value
* @param b buffer to which values are to be written
*/
-template
-void addSlice(const std::vector &s, gsl::span b) {
+template void addSlice(std::vector const& s, gsl::span b) {
addSlice(gsl::make_span(s.data(), s.size()), b);
};
@@ -131,21 +127,21 @@ void addSlice(const std::vector &s, gsl::span b) {
* @param s computed value
* @param b buffer to which values are to be written
*/
-void writeSlice(const AmiVector &s, gsl::span b);
-
+void writeSlice(AmiVector const& s, gsl::span b);
/**
- * @brief Remove parameter scaling according to the parameter scaling in pscale
- *
- * All vectors must be of same length.
- *
- * @param bufferScaled scaled parameters
- * @param pscale parameter scaling
- * @param bufferUnscaled unscaled parameters are written to the array
- */
-void unscaleParameters(gsl::span bufferScaled,
- gsl::span pscale,
- gsl::span bufferUnscaled);
+ * @brief Remove parameter scaling according to the parameter scaling in pscale
+ *
+ * All vectors must be of same length.
+ *
+ * @param bufferScaled scaled parameters
+ * @param pscale parameter scaling
+ * @param bufferUnscaled unscaled parameters are written to the array
+ */
+void unscaleParameters(
+ gsl::span bufferScaled,
+ gsl::span pscale, gsl::span bufferUnscaled
+);
/**
* @brief Remove parameter scaling according to `scaling`
@@ -173,16 +169,18 @@ double getScaledParameter(double unscaledParameter, ParameterScaling scaling);
* @param pscale parameter scaling
* @param bufferScaled destination
*/
-void scaleParameters(gsl::span bufferUnscaled,
- gsl::span pscale,
- gsl::span bufferScaled);
+void scaleParameters(
+ gsl::span bufferUnscaled,
+ gsl::span pscale, gsl::span bufferScaled
+);
/**
* @brief Returns the current backtrace as std::string
* @param maxFrames Number of frames to include
+ * @param first_frame Index of first frame to include
* @return Backtrace
*/
-std::string backtraceString(int maxFrames);
+std::string backtraceString(int maxFrames, int const first_frame = 0);
/**
* @brief Convert std::regex_constants::error_type to string
@@ -197,7 +195,7 @@ std::string regexErrorToString(std::regex_constants::error_type err_type);
* @param ap Argument list pointer
* @return Formatted String
*/
-std::string printfToString(const char *fmt, va_list ap);
+std::string printfToString(char const* fmt, va_list ap);
/**
* @brief Generic implementation for a context manager, explicitly deletes copy
diff --git a/include/amici/steadystateproblem.h b/include/amici/steadystateproblem.h
index 030ce7317e..bb1b39e36e 100644
--- a/include/amici/steadystateproblem.h
+++ b/include/amici/steadystateproblem.h
@@ -185,7 +185,6 @@ class SteadystateProblem {
/**
* @brief Handles the computation of quadratures in adjoint mode
- * @param newtonSolver pointer to the newtonSolver solver object
* @param solver pointer to the solver object
* @param model pointer to the model object
*/
diff --git a/python/examples/example_errors.ipynb b/python/examples/example_errors.ipynb
new file mode 100644
index 0000000000..d4f207c819
--- /dev/null
+++ b/python/examples/example_errors.ipynb
@@ -0,0 +1,1001 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "id": "6d8e8890",
+ "metadata": {},
+ "source": [
+ "# Debugging simulation failures\n",
+ "\n",
+ "**Objective:** Demonstrate common simulation failures and give some hints for interpreting, debugging, and fixing them."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "87383e84",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "%matplotlib inline\n",
+ "import os\n",
+ "import amici\n",
+ "from amici.petab_import import import_petab_problem\n",
+ "from amici.petab_objective import simulate_petab, RDATAS, EDATAS\n",
+ "from amici.plotting import plot_state_trajectories, plot_jacobian\n",
+ "import petab\n",
+ "import numpy as np\n",
+ "import matplotlib.pyplot as plt\n",
+ "from pathlib import Path\n",
+ "from contextlib import suppress\n",
+ "\n",
+ "try:\n",
+ " import benchmark_models_petab\n",
+ "except ModuleNotFoundError:\n",
+ " # install `benchmark_models_petab` if necessary\n",
+ " %pip install -q -e \"git+https://github.com/Benchmarking-Initiative/Benchmark-Models-PEtab.git@master#subdirectory=src/python&egg=benchmark_models_petab\"\n",
+ " try:\n",
+ " import benchmark_models_petab\n",
+ " except ModuleNotFoundError:\n",
+ " print(\"** Please restart the kernel. **\")"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "46500fb0",
+ "metadata": {},
+ "source": [
+ "## Overview\n",
+ "\n",
+ "In the following, we will simulate models contained in the [PEtab Benchmark Collection](https://github.com/Benchmarking-Initiative/Benchmark-Models-PEtab/) to demonstrate a number of simulation failures to analyze and fix them. We use the PEtab format, as it makes model import and simulation much easier, but everything illustrated here, also applies to plain SBML or PySB import.\n",
+ "\n",
+ "Note that, due to numerical issues, the examples below may not be fully reproducible on every system.\n",
+ "\n",
+ "If any simulation failures occur, they will be printed via Python logging. \n",
+ "\n",
+ "Programmatically, simulation success can be checked via `ReturnDataView.status`. In case of a successful simulation, and only then, this value corresponds to `amici.AMICI_SUCCESS`.\n",
+ "In case of a simulation error, all quantities in `ReturnData`/`ReturnDataView` will be reported up to the time of failure, the rest will be `NaN`. The likelihood and it's gradient will always be `NaN` in case of failure."
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "0ad882ac",
+ "metadata": {},
+ "source": [
+ "## `AMICI_TOO_MUCH_WORK` - `mxstep steps taken before reaching tout`\n",
+ "\n",
+ "Let's run a simulation:\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "ae3cb45e",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "petab_problem = benchmark_models_petab.get_problem(\"Fujita_SciSignal2010\")\n",
+ "amici_model = import_petab_problem(petab_problem, verbose=False, force_compile=False)\n",
+ "\n",
+ "np.random.seed(2991)\n",
+ "problem_parameters = dict(\n",
+ " zip(\n",
+ " petab_problem.x_free_ids,\n",
+ " petab_problem.sample_parameter_startpoints(n_starts=1)[0],\n",
+ " )\n",
+ ")\n",
+ "res = simulate_petab(\n",
+ " petab_problem=petab_problem, \n",
+ " amici_model=amici_model,\n",
+ " problem_parameters=problem_parameters,\n",
+ " scaled_parameters=True\n",
+ ")\n",
+ "print(\"Status:\", [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]])\n",
+ "assert [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]] == ['AMICI_SUCCESS', 'AMICI_SUCCESS', 'AMICI_SUCCESS', 'AMICI_TOO_MUCH_WORK', 'AMICI_SUCCESS', 'AMICI_SUCCESS']"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "e75a6dcc",
+ "metadata": {},
+ "source": [
+ "**What happened?**\n",
+ "\n",
+ "AMICI failed to integrate the forward problem. The problem occurred for only one simulation condition, `condition_step_03_0`. The issue occurred at $t = 3031.8$, where the CVODES reached the maximum number of steps.\n",
+ "\n",
+ "**How to address?**\n",
+ "\n",
+ "The number of steps the solver has to take is closely related to the chosen error tolerance. More accurate results, more steps. Therefore, this problem can be solved in two ways:\n",
+ "\n",
+ "1. Increasing the maximum number of steps via [amici.Solver.setMaxSteps](https://amici.readthedocs.io/en/latest/generated/amici.amici.Solver.html#amici.amici.Solver.setMaxSteps). Note that this will increase the time required for simulation, and that simulation may still fail eventually. Sometimes it may be preferable to not increase this limit but rather fail fast. Also note that increasing the number of allowed steps increase RAM requirements (even if fewer steps are actually taken), so don't set this to ridiculously large values in order to avoid this error.\n",
+ "\n",
+ "2. Reducing the number of steps CVODES has to take. This is determined by the required error tolerance. There are various solver error tolerances than can be adjusted. The most relevant ones are those controlled via [amici.Solver.setRelativeTolerance()](https://amici.readthedocs.io/en/latest/generated/amici.amici.Solver.html#amici.amici.Solver.setRelativeTolerance) and [amici.Solver.setAbsoluteTolerance()](https://amici.readthedocs.io/en/latest/generated/amici.amici.Solver.html#amici.amici.Solver.setAbsoluteTolerance).\n",
+ "\n",
+ "So, let's fix that:"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "e9667f55",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# let's increase the allowed number of steps by 10x:\n",
+ "print(\"Increasing allowed number of steps ...\")\n",
+ "amici_solver = amici_model.getSolver()\n",
+ "amici_solver.setMaxSteps(10 * amici_solver.getMaxSteps())\n",
+ "\n",
+ "res = simulate_petab(\n",
+ " petab_problem=petab_problem, \n",
+ " amici_model=amici_model,\n",
+ " problem_parameters=problem_parameters,\n",
+ " scaled_parameters=True,\n",
+ " solver=amici_solver\n",
+ ")\n",
+ "\n",
+ "print(\"Status:\", [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]])\n",
+ "assert all(rdata.status == amici.AMICI_SUCCESS for rdata in res[RDATAS])\n",
+ "print(\"Simulations finished succesfully.\")\n",
+ "print()\n",
+ "\n",
+ "\n",
+ "# let's relax the relative error tolerance by a factor of 50\n",
+ "print(\"Relaxing relative error tolerance ...\")\n",
+ "amici_solver = amici_model.getSolver()\n",
+ "amici_solver.setRelativeTolerance(50 * amici_solver.getRelativeTolerance())\n",
+ "\n",
+ "res = simulate_petab(\n",
+ " petab_problem=petab_problem, \n",
+ " amici_model=amici_model,\n",
+ " problem_parameters=problem_parameters,\n",
+ " scaled_parameters=True,\n",
+ " solver=amici_solver\n",
+ ")\n",
+ "print(\"Status:\", [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]])\n",
+ "assert all(rdata.status == amici.AMICI_SUCCESS for rdata in res[RDATAS])\n",
+ "print(\"Simulations finished succesfully.\")\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "18fd3fa6",
+ "metadata": {},
+ "source": [
+ "## `Internal t = [...] and h = [...] are such that t + h = t on the next step`\n",
+ "\n",
+ "Let's run a simulation:"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "f78179dc",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "petab_problem = benchmark_models_petab.get_problem(\"Crauste_CellSystems2017\")\n",
+ "amici_model = import_petab_problem(petab_problem, verbose=False)\n",
+ "\n",
+ "np.random.seed(1)\n",
+ "problem_parameters = dict(\n",
+ " zip(\n",
+ " petab_problem.x_free_ids,\n",
+ " petab_problem.sample_parameter_startpoints(n_starts=1)[0],\n",
+ " )\n",
+ ")\n",
+ "res = simulate_petab(\n",
+ " petab_problem=petab_problem, \n",
+ " amici_model=amici_model,\n",
+ " problem_parameters=problem_parameters,\n",
+ " scaled_parameters=True\n",
+ ")\n",
+ "print(\"Status:\", [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]])\n",
+ "assert [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]] == ['AMICI_TOO_MUCH_WORK']"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "53e4b822",
+ "metadata": {},
+ "source": [
+ "**What happened?**\n",
+ "\n",
+ "The forward simulation failed because AMICI the solver exceeded the maximum number of steps. Unlike in the previous case of `mxstep steps taken before reaching tout` (see above), here we got several additional warnings that the current step size $h$ is numerically zero.\n",
+ "\n",
+ "**How to address?**\n",
+ "\n",
+ "The warning `Internal t = [...] and h = [...] are such that t + h = t on the next step` tells us that the solver is not able to move forward. The solver may be able to recover from that, but not always.\n",
+ "\n",
+ "Let's look at the state trajectories to see what's going on. Such a tiny step size is usually related to very fast dynamics. We repeat the simulation with additional timepoints before the point of failure:"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "6a6794d3",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# Create a copy of this simulation condition\n",
+ "edata = amici.ExpData(res[EDATAS][0])\n",
+ "edata.setTimepoints(np.linspace(0, 0.33011, 5000))\n",
+ "amici_solver = amici_model.getSolver()\n",
+ "rdata = amici.runAmiciSimulation(amici_model, amici_solver, edata)\n",
+ "\n",
+ "# Visualize state trajectories\n",
+ "plot_state_trajectories(rdata, model=amici_model)\n",
+ "plt.yscale(\"log\")"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "a7fe6d08",
+ "metadata": {},
+ "source": [
+ "We can see a steep increase for `Pathogen` just before the error occurs. Let's zoom in:"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "6a645efb",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "plt.plot(rdata.t, rdata.by_id(\"Pathogen\"))\n",
+ "plt.xlabel(\"time\")\n",
+ "plt.ylabel(\"Pathogen\");"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "b85cd813",
+ "metadata": {},
+ "source": [
+ "The solver is unable to handle such a steep increase. There is not much we can do. Increasing the tolerances will let the solver proceed a bit further, but this is usually not enough. Most likely there is a problem in the model or in the choice of parameter values."
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "5afbd242",
+ "metadata": {},
+ "source": [
+ "## `the error test failed repeatedly or with |h| = hmin`\n",
+ "\n",
+ "Let's run a simulation:"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "bb8910f2",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "petab_problem = benchmark_models_petab.get_problem(\"Fujita_SciSignal2010\")\n",
+ "amici_model = import_petab_problem(petab_problem, verbose=False)\n",
+ "\n",
+ "np.random.seed(4920)\n",
+ "problem_parameters = dict(\n",
+ " zip(\n",
+ " petab_problem.x_free_ids,\n",
+ " petab_problem.sample_parameter_startpoints(n_starts=1)[0],\n",
+ " )\n",
+ ")\n",
+ "res = simulate_petab(\n",
+ " petab_problem=petab_problem, \n",
+ " amici_model=amici_model,\n",
+ " problem_parameters=problem_parameters,\n",
+ " scaled_parameters=True\n",
+ ")\n",
+ "print(\"Status:\", [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]])\n",
+ "\n",
+ "assert [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]] == ['AMICI_SUCCESS', 'AMICI_ERR_FAILURE', 'AMICI_SUCCESS', 'AMICI_SUCCESS', 'AMICI_SUCCESS', 'AMICI_SUCCESS']"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "0b3a9904",
+ "metadata": {},
+ "source": [
+ "**What happened?**\n",
+ "\n",
+ "AMICI failed to integrate the forward problem. The problem occurred for only one simulation condition, `condition_step_00_3`. The issue occurred at $t = 429.232$, where the error test failed.\n",
+ "This means, the solver is unable to take a step of non-zero size without violating the choosen error tolerances."
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "c16ac6c8",
+ "metadata": {},
+ "source": [
+ "**How to address?**\n",
+ "\n",
+ "The step size is computed based on the Jacobian. Inspecting `ReturnData.J` shows us that we have rather large values in the Jacobian:"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "467c3d36",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "rdata = res[RDATAS][1]\n",
+ "\n",
+ "# Show Jacobian as heatmap\n",
+ "plot_jacobian(rdata)\n",
+ "\n",
+ "print(f\"largest absolute Jacobian value: {np.max(np.abs(rdata.J)):.3g}\")"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "1f6ec0e3",
+ "metadata": {},
+ "source": [
+ "In this case, the default relative error tolerance may be too high and lead too large absolute errors. \n",
+ "\n",
+ "Let's retry simulation using stricter tolerances:"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "600ae826",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# set stricter relative error tolerance\n",
+ "amici_solver = amici_model.getSolver()\n",
+ "amici_solver.setRelativeTolerance(amici_solver.getRelativeTolerance() / 10)\n",
+ "\n",
+ "res = simulate_petab(\n",
+ " petab_problem=petab_problem, \n",
+ " amici_model=amici_model,\n",
+ " problem_parameters=problem_parameters,\n",
+ " scaled_parameters=True,\n",
+ " solver=amici_solver\n",
+ ")\n",
+ "print(\"Status:\", [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]])\n",
+ "assert all(rdata.status == amici.AMICI_SUCCESS for rdata in res[RDATAS])\n",
+ "print(\"Simulations finished succesfully.\")"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "616710b6",
+ "metadata": {},
+ "source": [
+ "## `Cvode routine CVode returned a root after reinitialization`\n",
+ "\n",
+ "Let's run a simulation:"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "cec31332",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "petab_problem = benchmark_models_petab.get_problem(\"Weber_BMC2015\")\n",
+ "amici_model = import_petab_problem(petab_problem, verbose=False, force_compile=False)\n",
+ "\n",
+ "np.random.seed(4)\n",
+ "problem_parameters = dict(\n",
+ " zip(\n",
+ " petab_problem.x_free_ids,\n",
+ " petab_problem.sample_parameter_startpoints(n_starts=1)[0],\n",
+ " )\n",
+ ")\n",
+ "res = simulate_petab(\n",
+ " petab_problem=petab_problem, \n",
+ " amici_model=amici_model,\n",
+ " problem_parameters=problem_parameters,\n",
+ " scaled_parameters=True\n",
+ ")\n",
+ "print(\"Status:\", [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]])\n",
+ "assert [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]] == ['AMICI_ERROR', 'AMICI_SUCCESS']"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "d4386603",
+ "metadata": {},
+ "source": [
+ "**What happened?**\n",
+ "\n",
+ "The simulation failed because the initial step-size after an event or heaviside function was too small. The error occured during simulation of condition `model1_data1` after successful preequilibration (`model1_data2`)."
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "02f2dd5d",
+ "metadata": {},
+ "source": [
+ "**How to address?**\n",
+ "\n",
+ "The error message already suggests a fix for this situation, so let's try increasing the relative tolerance:"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "3d1552e3",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "amici_solver = amici_model.getSolver()\n",
+ "amici_solver.setRelativeTolerance(200 * amici_solver.getRelativeTolerance())\n",
+ "\n",
+ "np.random.seed(4)\n",
+ "problem_parameters = dict(\n",
+ " zip(\n",
+ " petab_problem.x_free_ids,\n",
+ " petab_problem.sample_parameter_startpoints(n_starts=1)[0],\n",
+ " )\n",
+ ")\n",
+ "res = simulate_petab(\n",
+ " petab_problem=petab_problem, \n",
+ " amici_model=amici_model,\n",
+ " problem_parameters=problem_parameters,\n",
+ " scaled_parameters=True,\n",
+ " solver=amici_solver\n",
+ ")\n",
+ "print(\"Status:\", [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]])\n",
+ "assert all(rdata.status == amici.AMICI_SUCCESS for rdata in res[RDATAS])"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "86f4db29",
+ "metadata": {},
+ "source": [
+ "## `AMICI encountered a NaN / Inf value for [...]`\n",
+ "\n",
+ "Let's run a simulation:"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "d97349ff",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "petab_problem = benchmark_models_petab.get_problem(\"Borghans_BiophysChem1997\")\n",
+ "amici_model = import_petab_problem(petab_problem, verbose=False)\n",
+ "\n",
+ "np.random.seed(18)\n",
+ "problem_parameters = dict(\n",
+ " zip(\n",
+ " petab_problem.x_free_ids,\n",
+ " petab_problem.sample_parameter_startpoints(n_starts=1)[0],\n",
+ " )\n",
+ ")\n",
+ "res = simulate_petab(\n",
+ " petab_problem=petab_problem, \n",
+ " amici_model=amici_model,\n",
+ " problem_parameters=problem_parameters,\n",
+ " scaled_parameters=True\n",
+ ")\n",
+ "print(\"Status:\", [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]])\n",
+ "assert [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]] == ['AMICI_FIRST_RHSFUNC_ERR']"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "63641cff",
+ "metadata": {},
+ "source": [
+ "**What happened?**\n",
+ "\n",
+ "The forward simulation failed because AMICI encountered a `NaN` value when simulating condition `model1_data1`.\n",
+ "Then `NaN`s occurred in $\\dot x$ and $w$ (model expressions, such as reaction fluxes or assignment rules). Furthermore, the failure occurred at the first call, so at $t = t_0$ (here: $t = 0$).\n",
+ "\n",
+ "**How to address?**\n",
+ "\n",
+ "The `NaN` in $\\dot x$ is most likely a consequence of the one in $w$. (A subset of) the dependency tree looks something like:\n",
+ "\n",
+ "[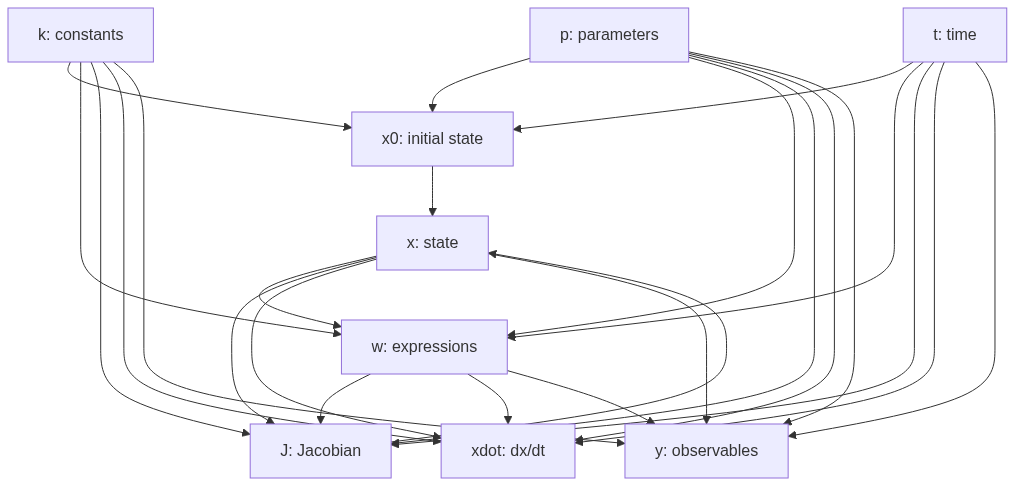](https://mermaid.live/edit#pako:eNpdkrFuwyAQhl8FIWVL1MwMndKFtd1wBmJIg2IDwucCivLuxQp2zvaA_P1398Od7kFbpzRl9DdIfyM_p8aS8t3FnZHW2QGkheH8Er3wjHgZZK9Bh1kFAYyA6XXldBTpyIixBozsSHGAJSQSWwlccEa4bN3FSFu1KCIjOvmgh8GUF8y1yoGYDkZU-lBQ5SwyI-4y6PAnL52esl-B3a68pDZDDofPhfyKYEVLacSVERdGXFchzYAu59iBYweOHTh2qBAxvLupI3s9eJ41pndqGdOqvYXThv2G7xuOiBf7jHMzNsr41oyvzNgv0z3tdeilUWXzHlOooXDTvW4oK79KX-XYQUMb-yypcgT3nW1LGYRR7-noVVmhk5FlZ3vKrrIbFvVLGXChis9_j9jNUw)\n",
+ "\n",
+ "Always look for the most basic (furthest up) model quantities.\n",
+ "In cases where there non-finite values occur in expressions further down, rerunning the simulation after calling `Model.setAlwaysCheckFinite(True)` may give some further hints on where the issue originates.\n",
+ "\n",
+ "The `NaN` in $w$ occurred for `flux_v7_v_6` (see error log), i.e., when computing the reaction flux for reaction `v7_v_6`. As $w$ only depends on $(t, p, k, x)$ and no non-finite values have been reported for those, the issue has to be in the respective flux equation.\n",
+ "\n",
+ "Let's look at that expression. This can either be done by inspecting the underlying SBML model (e.g., using COPASI), or by checking the generated model code:"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "fca95143",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# model name and source code location\n",
+ "model_name = amici_model.module.__package__\n",
+ "model_src_dir = Path(amici_model.module.__file__).parents[1]\n",
+ "\n",
+ "# find the problematic expression in the model source code\n",
+ "!grep flux_v7_v_6 {model_src_dir}/{model_name}_w.cpp"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "9f49a00a",
+ "metadata": {},
+ "source": [
+ "What could go wrong? We can obtain `NaN` from any of these symbols symbols being `NaN`, or through division by zero.\n",
+ "\n",
+ "Let's let's check the denominator first: $$(A\\_state^2 + Kp^2)*(Kd^{n\\_par} + Z\\_state^{n\\_par})$$\n",
+ "\n",
+ "\n",
+ "`A_state` and `Z_state` are state variables, `Kd`, `K_p`, and `n_par` are parameters.\n",
+ "\n",
+ "As the error occurred at $t = t_0$, let's ensure the initial state is non-zero and finite:"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "0eec6fe8",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "rdata = res[RDATAS][0]\n",
+ "edata = res[EDATAS][0]\n",
+ "# check initial states\n",
+ "x0 = dict(zip(amici_model.getStateIds(), rdata.x0))\n",
+ "print(f\"{x0=}\")"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "050c4c38",
+ "metadata": {},
+ "source": [
+ "The initial states are fine - the first multiplicand is non-zero, as $x_0$ was non-zero. \n",
+ "\n",
+ "So let's check the parameter values occurring in the second multiplicand:"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "ec724ce9",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# we have to account for the chosen parameter scale\n",
+ "from itertools import starmap\n",
+ "unscaled_parameter = dict(zip(\n",
+ " amici_model.getParameterIds(),\n",
+ " starmap(amici.getUnscaledParameter, zip(edata.parameters, edata.pscale)),\n",
+ "))\n",
+ "print(dict((p, unscaled_parameter[p]) for p in ('Kd', 'Kp', 'n_par')))"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "62d82971",
+ "metadata": {},
+ "source": [
+ "Considering that `n_par` occurrs as exponent, it's magnitude looks pretty high.\n",
+ "This term is very likely causing the problem - let's check:"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "9f3f8bdb",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "print(f\"{x0['Z_state']**unscaled_parameter['n_par'] + unscaled_parameter['Kd']**unscaled_parameter['n_par']=}\")"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "6616c2ff",
+ "metadata": {},
+ "source": [
+ "Indeed, no way we can fix this for the given model.\n",
+ "This was most likely an unrealistic parameter value, originating from a too high upper parameter bound for `n_par`.\n",
+ "Therefore, if this error occurs during optimization, a first step could be adapting the respective parameter bounds.\n",
+ "In other cases, this may be a result of unfortunate arrangement of model expressions, which can sometimes be solved by passing a suitable simplification function to the model import."
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "22cfbbbf",
+ "metadata": {},
+ "source": [
+ "\n",
+ "\n",
+ "## `Steady state sensitivity computation failed due to unsuccessful factorization of RHS Jacobian`\n",
+ "\n",
+ "Let's run a simulation:"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "b41a5017",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "petab_problem = benchmark_models_petab.get_problem(\"Blasi_CellSystems2016\")\n",
+ "with suppress(KeyError):\n",
+ " del os.environ[\"AMICI_EXPERIMENTAL_SBML_NONCONST_CLS\"]\n",
+ "amici_model = import_petab_problem(\n",
+ " petab_problem, \n",
+ " verbose=False,\n",
+ " force_compile=True,\n",
+ " model_name=\"Blasi_CellSystems2016_1\"\n",
+ ")\n",
+ "\n",
+ "amici_solver = amici_model.getSolver()\n",
+ "amici_solver.setSensitivityMethod(amici.SensitivityMethod.forward)\n",
+ "amici_solver.setSensitivityOrder(amici.SensitivityOrder.first)\n",
+ "\n",
+ "np.random.seed(2020)\n",
+ "problem_parameters = dict(\n",
+ " zip(\n",
+ " petab_problem.x_free_ids,\n",
+ " petab_problem.sample_parameter_startpoints(n_starts=1)[0],\n",
+ " )\n",
+ ")\n",
+ "res = simulate_petab(\n",
+ " petab_problem=petab_problem, \n",
+ " amici_model=amici_model,\n",
+ " problem_parameters=problem_parameters,\n",
+ " scaled_parameters=True,\n",
+ " solver=amici_solver,\n",
+ ")\n",
+ "print(\"Status:\", [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]])\n",
+ "\n",
+ "# hard to reproduce on GHA\n",
+ "if os.getenv('GITHUB_ACTIONS') is None:\n",
+ " assert [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]] == ['AMICI_ERROR']"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "82267e47",
+ "metadata": {},
+ "source": [
+ "**What happened?**\n",
+ "\n",
+ "AMICI failed to compute steadystate sensitivities, because it was not able to factorize the Jacobian.\n",
+ "\n",
+ "**How to address?**\n",
+ "\n",
+ "This is most likely a result of a singular Jacobian. Let's check the condition number:"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "8cd349b7",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "rdata = res[RDATAS][0]\n",
+ "np.linalg.cond(rdata.J)"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "da131d3e",
+ "metadata": {},
+ "source": [
+ "Indeed, the condition number shows that the Jacobian is numerically singular. If this happens consistently, it is usually due to conserved quantities in the model.\n",
+ "\n",
+ "There are two ways we can address that:\n",
+ "\n",
+ "1. Use numerical integration to compute sensitivities, for which a singular Jacobian is not an issue. This is, usually, slower, though.\n",
+ "2. Remove any conserved quantities.\n",
+ "\n",
+ "Let's try both approaches:"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "f82078e7",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# use numerical integration\n",
+ "amici_model.setSteadyStateSensitivityMode(amici.SteadyStateSensitivityMode.integrationOnly)\n",
+ "\n",
+ "res = simulate_petab(\n",
+ " petab_problem=petab_problem, \n",
+ " amici_model=amici_model,\n",
+ " problem_parameters=problem_parameters,\n",
+ " scaled_parameters=True,\n",
+ " solver=amici_solver,\n",
+ ")\n",
+ "print(\"Status:\", [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]])\n",
+ "assert all(rdata.status == amici.AMICI_SUCCESS for rdata in res[RDATAS])"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "8d7be541",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# Remove conserved quantities - this requires re-importing the model\n",
+ "\n",
+ "# this is enabled by the `AMICI_EXPERIMENTAL_SBML_NONCONST_CLS` environment variable\n",
+ "os.environ[\"AMICI_EXPERIMENTAL_SBML_NONCONST_CLS\"] = \"1\"\n",
+ "amici_model = import_petab_problem(\n",
+ " petab_problem, \n",
+ " verbose=False,\n",
+ " # we need a different model name if we import the model again\n",
+ " # we cannot load a model with the same name as an already loaded model\n",
+ " model_name=\"Blasi_CellSystems2016_2\",\n",
+ " force_compile=True,\n",
+ ")\n",
+ "del os.environ[\"AMICI_EXPERIMENTAL_SBML_NONCONST_CLS\"]\n",
+ "\n",
+ "amici_solver = amici_model.getSolver()\n",
+ "amici_solver.setSensitivityMethod(amici.SensitivityMethod.forward)\n",
+ "amici_solver.setSensitivityOrder(amici.SensitivityOrder.first)\n",
+ "\n",
+ "res = simulate_petab(\n",
+ " petab_problem=petab_problem, \n",
+ " amici_model=amici_model,\n",
+ " problem_parameters=problem_parameters,\n",
+ " scaled_parameters=True,\n",
+ " solver=amici_solver,\n",
+ ")\n",
+ "print(\"Status:\", [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]])\n",
+ "assert all(rdata.status == amici.AMICI_SUCCESS for rdata in res[RDATAS])"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "4b977c0b",
+ "metadata": {},
+ "source": [
+ "## `Steady state computation failed`\n",
+ "\n",
+ "Let's run a simulation:"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "97f797dd",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "petab_problem = benchmark_models_petab.get_problem(\"Brannmark_JBC2010\")\n",
+ "amici_model = import_petab_problem(\n",
+ " petab_problem, \n",
+ " verbose=False,\n",
+ ")\n",
+ "\n",
+ "amici_solver = amici_model.getSolver()\n",
+ "\n",
+ "\n",
+ "np.random.seed(1851)\n",
+ "problem_parameters = dict(\n",
+ " zip(\n",
+ " petab_problem.x_free_ids,\n",
+ " petab_problem.sample_parameter_startpoints(n_starts=1)[0],\n",
+ " )\n",
+ ")\n",
+ "res = simulate_petab(\n",
+ " petab_problem=petab_problem, \n",
+ " amici_model=amici_model,\n",
+ " problem_parameters=problem_parameters,\n",
+ " scaled_parameters=True,\n",
+ " solver=amici_solver,\n",
+ ")\n",
+ " \n",
+ "print(\"Status:\", [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]])\n",
+ "\n",
+ "# hard to reproduce on GHA\n",
+ "if os.getenv('GITHUB_ACTIONS') is None:\n",
+ " assert [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]] == ['AMICI_ERROR', 'AMICI_SUCCESS', 'AMICI_SUCCESS', 'AMICI_SUCCESS', 'AMICI_SUCCESS', 'AMICI_SUCCESS', 'AMICI_SUCCESS', 'AMICI_SUCCESS']"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "0713b830",
+ "metadata": {},
+ "source": [
+ "**What happened?**\n",
+ "\n",
+ "All given experimental conditions require pre-equilibration, i.e., finding a steady state. AMICI first tries find a steady state using the Newton solver, if that fails, it tries simulating until steady state, if that also failes, it tries the Newton solver from the end of the simulation. In this case, all three failed. Neither Newton's method nor simulation yielded a steadystate satisfying the required tolerances.\n",
+ "\n",
+ "This can also be seen in `ReturnDataView.preeq_status` (the three statuses corresponds to Newton \\#1, Simulation, Newton \\#2):"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "ffdc8e82",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "rdata = res[RDATAS][0]\n",
+ "list(map(amici.SteadyStateStatus, rdata.preeq_status.flatten()))"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "189dd964",
+ "metadata": {},
+ "source": [
+ "**How to address?**\n",
+ "\n",
+ "There are several ways to address that:\n",
+ "\n",
+ "1. Stricter integration tolerances (preferred if affordable - higher accuracy, but generally slower)\n",
+ "\n",
+ "2. Looser steadystate tolerances (lower accuracy, generally faster)\n",
+ "\n",
+ "3. Increase the number of allowed steps for Newton's method\n",
+ "\n",
+ "Let's try all of them:"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "28fada9f",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# Reduce relative tolerance for integration\n",
+ "amici_solver = amici_model.getSolver()\n",
+ "amici_solver.setRelativeTolerance(1/100 * amici_solver.getRelativeTolerance())\n",
+ "\n",
+ "res = simulate_petab(\n",
+ " petab_problem=petab_problem, \n",
+ " amici_model=amici_model,\n",
+ " problem_parameters=problem_parameters,\n",
+ " scaled_parameters=True,\n",
+ " solver=amici_solver,\n",
+ ")\n",
+ "print(\"status:\", [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]])\n",
+ "\n",
+ "rdata = res[RDATAS][0]\n",
+ "print(f\"preeq_status={list(map(amici.SteadyStateStatus, rdata.preeq_status.flatten()))}\")\n",
+ "print(f\"{rdata.preeq_numsteps=}\")\n",
+ "\n",
+ "# hard to reproduce on GHA\n",
+ "if os.getenv('GITHUB_ACTIONS') is None:\n",
+ " assert all(rdata.status == amici.AMICI_SUCCESS for rdata in res[RDATAS])"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "a6d34467",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# Increase relative steady state tolerance\n",
+ "for log10_relaxation_factor in range(1, 10):\n",
+ " print(f\"Relaxing tolerances by factor {10 ** log10_relaxation_factor}\")\n",
+ " amici_solver = amici_model.getSolver()\n",
+ " amici_solver.setRelativeToleranceSteadyState(amici_solver.getRelativeToleranceSteadyState() * 10 ** log10_relaxation_factor)\n",
+ " \n",
+ " res = simulate_petab(\n",
+ " petab_problem=petab_problem, \n",
+ " amici_model=amici_model,\n",
+ " problem_parameters=problem_parameters,\n",
+ " scaled_parameters=True,\n",
+ " solver=amici_solver,\n",
+ " )\n",
+ " if all(rdata.status == amici.AMICI_SUCCESS for rdata in res[RDATAS]):\n",
+ " print(f\"-> Succeeded with relative steady state tolerance {amici_solver.getRelativeToleranceSteadyState()}\\n\")\n",
+ " break\n",
+ " else:\n",
+ " print(\"-> Failed.\\n\")\n",
+ "\n",
+ "print(\"status:\", [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]])\n",
+ "\n",
+ "rdata = res[RDATAS][0]\n",
+ "print(f\"preeq_status={list(map(amici.SteadyStateStatus, rdata.preeq_status.flatten()))}\")\n",
+ "print(f\"{rdata.preeq_numsteps=}\")\n",
+ "assert all(rdata.status == amici.AMICI_SUCCESS for rdata in res[RDATAS])"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "6d1b1835",
+ "metadata": {},
+ "source": [
+ "That fixed the error, and took only a quarter of the number steps as the previous run, but at the cost of much lower accuracy."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "df1ee3fc",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# Let's try increasing the number of Newton steps\n",
+ "# (this is 0 by default, so the Newton solver wasn't used before, \n",
+ "# as can be seen from the 0 in `rdata.preeq_numsteps[0]`)\n",
+ "amici_solver = amici_model.getSolver()\n",
+ "amici_solver.setNewtonMaxSteps(10**4)\n",
+ "\n",
+ "res = simulate_petab(\n",
+ " petab_problem=petab_problem, \n",
+ " amici_model=amici_model,\n",
+ " problem_parameters=problem_parameters,\n",
+ " scaled_parameters=True,\n",
+ " solver=amici_solver,\n",
+ ")\n",
+ "print(\"status:\", [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]])\n",
+ "\n",
+ "rdata = res[RDATAS][0]\n",
+ "print(f\"preeq_status={list(map(amici.SteadyStateStatus, rdata.preeq_status.flatten()))}\")\n",
+ "print(f\"{rdata.preeq_numsteps=}\")\n",
+ "# hard to reproduce on GHA\n",
+ "if os.getenv('GITHUB_ACTIONS') is None:\n",
+ " assert [amici.simulation_status_to_str(rdata.status) for rdata in res[RDATAS]] == ['AMICI_ERROR', 'AMICI_SUCCESS', 'AMICI_SUCCESS', 'AMICI_SUCCESS', 'AMICI_SUCCESS', 'AMICI_SUCCESS', 'AMICI_SUCCESS', 'AMICI_SUCCESS']"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "5c746982",
+ "metadata": {},
+ "source": [
+ "Increasing the maximum number of Newton steps doesn't seem to help here. The Jacobian was numerically singular and its factorization failed. This can be a result of conserved quantities in the model. Section [Steady state sensitivity computation failed due to unsuccessful factorization of RHS Jacobian](#unsuccessful_factorization) shows how to address that."
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "Python 3",
+ "language": "python",
+ "name": "python3"
+ },
+ "language_info": {
+ "codemirror_mode": {
+ "name": "ipython",
+ "version": 3
+ },
+ "file_extension": ".py",
+ "mimetype": "text/x-python",
+ "name": "python",
+ "nbconvert_exporter": "python",
+ "pygments_lexer": "ipython3",
+ "version": "3.10.6"
+ },
+ "toc": {
+ "base_numbering": 1,
+ "nav_menu": {},
+ "number_sections": true,
+ "sideBar": true,
+ "skip_h1_title": false,
+ "title_cell": "Table of Contents",
+ "title_sidebar": "Contents",
+ "toc_cell": false,
+ "toc_position": {
+ "height": "calc(100% - 180px)",
+ "left": "10px",
+ "top": "150px",
+ "width": "335.6px"
+ },
+ "toc_section_display": true,
+ "toc_window_display": true
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 5
+}
diff --git a/python/sdist/amici/__init__.py b/python/sdist/amici/__init__.py
index 94acc73df1..a55bfd5115 100644
--- a/python/sdist/amici/__init__.py
+++ b/python/sdist/amici/__init__.py
@@ -120,14 +120,19 @@ def _imported_from_setup() -> bool:
from .sbml_import import SbmlImporter, assignmentRules2observables
from .ode_export import ODEModel, ODEExporter
- from typing import Protocol
+ from typing import Protocol, runtime_checkable
+ @runtime_checkable
class ModelModule(Protocol):
"""Enable Python static type checking for AMICI-generated model
modules"""
+
def getModel(self) -> amici.Model:
- pass
+ ...
+
+ def get_model(self) -> amici.Model:
+ ...
class add_path:
diff --git a/python/sdist/amici/__init__.template.py b/python/sdist/amici/__init__.template.py
index 9fbab85003..bc83c1d4c9 100644
--- a/python/sdist/amici/__init__.template.py
+++ b/python/sdist/amici/__init__.template.py
@@ -14,6 +14,7 @@
'version currently installed.'
)
-from TPL_MODELNAME._TPL_MODELNAME import *
+from .TPL_MODELNAME import *
+from .TPL_MODELNAME import getModel as get_model
__version__ = 'TPL_PACKAGE_VERSION'
diff --git a/python/sdist/amici/logging.py b/python/sdist/amici/logging.py
index eae753b29e..f115a1b2e7 100644
--- a/python/sdist/amici/logging.py
+++ b/python/sdist/amici/logging.py
@@ -187,16 +187,19 @@ def wrapper_timer(*args, **kwargs):
)
recursion = ''
+ level = logging.INFO
if recursion_level > 1:
recursion = '+' * (recursion_level - 1)
+ level = logging.DEBUG
tstart = time.perf_counter()
rval = func(*args, **kwargs)
tend = time.perf_counter()
spacers = ' ' * max(54 - len(description) - len(logger.name) -
len(recursion), 0)
- logger.info(f'Finished {description}{spacers}'
- f'{recursion} ({(tend - tstart):.2E}s)')
+ logger.log(
+ level, f'Finished {description}{spacers}{recursion} ({(tend - tstart):.2E}s)'
+ )
return rval
return wrapper_timer
return decorator_timer
diff --git a/python/sdist/amici/numpy.py b/python/sdist/amici/numpy.py
index 65df16a557..ce767679f3 100644
--- a/python/sdist/amici/numpy.py
+++ b/python/sdist/amici/numpy.py
@@ -141,6 +141,14 @@ def __deepcopy__(self, memo):
other._cache = copy.deepcopy(self._cache)
return other
+ def __repr__(self):
+ """
+ String representation of the object
+
+ :returns: string representation
+ """
+ return f'<{self.__class__.__name__}({self._swigptr})>'
+
class ReturnDataView(SwigPtrView):
"""
@@ -235,9 +243,13 @@ def __getitem__(self, item: str) -> Union[np.ndarray, ReturnDataPtr,
:returns: self[item]
"""
+ if item == 'status':
+ return int(super().__getitem__(item))
+
if item == 't':
item = 'ts'
- return super(ReturnDataView, self).__getitem__(item)
+
+ return super().__getitem__(item)
def by_id(
self,
diff --git a/python/sdist/amici/ode_export.py b/python/sdist/amici/ode_export.py
index fcae4d883c..bc669374af 100644
--- a/python/sdist/amici/ode_export.py
+++ b/python/sdist/amici/ode_export.py
@@ -1872,7 +1872,7 @@ def _derivative(self, eq: str, var: str, name: str = None) -> None:
cv for cv in ['w', 'tcl']
if var_in_function_signature(eq, cv)
and cv not in self._lock_total_derivative
- and var is not cv
+ and var != cv
and min(self.sym(cv).shape)
and (
(eq, var) not in ignore_chainrule
@@ -3070,7 +3070,7 @@ def _write_model_header_cpp(self) -> None:
self._get_symbol_name_initializer_list('y'),
'OBSERVABLE_TRAFO_INITIALIZER_LIST':
'\n'.join(
- f'ObservableScaling::{trafo}, // y[{idx}]'
+ f'ObservableScaling::{trafo.value}, // y[{idx}]'
for idx, trafo in enumerate(
self.model.get_observable_transformations()
)
diff --git a/python/sdist/amici/ode_model.py b/python/sdist/amici/ode_model.py
index a21526d27d..9e03822ac3 100644
--- a/python/sdist/amici/ode_model.py
+++ b/python/sdist/amici/ode_model.py
@@ -297,12 +297,15 @@ class Observable(ModelQuantity):
_measurement_symbol: Union[sp.Symbol, None] = None
- def __init__(self,
- identifier: sp.Symbol,
- name: str,
- value: sp.Expr,
- measurement_symbol: Optional[sp.Symbol] = None,
- transformation: Optional[ObservableTransformation] = 'lin'):
+ def __init__(
+ self,
+ identifier: sp.Symbol,
+ name: str,
+ value: sp.Expr,
+ measurement_symbol: Optional[sp.Symbol] = None,
+ transformation: Optional[
+ ObservableTransformation] = ObservableTransformation.LIN
+ ):
"""
Create a new Observable instance.
diff --git a/python/sdist/amici/petab_objective.py b/python/sdist/amici/petab_objective.py
index e8557cb242..4f03e66647 100644
--- a/python/sdist/amici/petab_objective.py
+++ b/python/sdist/amici/petab_objective.py
@@ -37,6 +37,7 @@
RES = 'res'
SRES = 'sres'
RDATAS = 'rdatas'
+EDATAS = 'edatas'
@log_execution_time('Simulating PEtab model', logger)
@@ -92,6 +93,7 @@ def simulate_petab(
* cost function value (``LLH``),
* list of :class:`amici.amici.ReturnData` (``RDATAS``),
+ * list of :class:`amici.amici.ExpData` (``EDATAS``),
corresponding to the different simulation conditions.
For ordering of simulation conditions, see
@@ -163,7 +165,8 @@ def simulate_petab(
return {
LLH: llh,
- RDATAS: rdatas
+ RDATAS: rdatas,
+ EDATAS: edatas,
}
diff --git a/python/sdist/amici/plotting.py b/python/sdist/amici/plotting.py
index d2917de9fe..6b3eade225 100644
--- a/python/sdist/amici/plotting.py
+++ b/python/sdist/amici/plotting.py
@@ -3,14 +3,17 @@
--------
Plotting related functions
"""
-from . import ReturnDataView, Model
+from typing import Iterable, Optional
import matplotlib.pyplot as plt
+import pandas as pd
+import seaborn as sns
from matplotlib.axes import Axes
-from typing import Optional, Iterable
+from . import Model, ReturnDataView
-def plotStateTrajectories(
+
+def plot_state_trajectories(
rdata: ReturnDataView,
state_indices: Optional[Iterable[int]] = None,
ax: Optional[Axes] = None,
@@ -50,7 +53,7 @@ def plotStateTrajectories(
ax.set_title('State trajectories')
-def plotObservableTrajectories(
+def plot_observable_trajectories(
rdata: ReturnDataView,
observable_indices: Optional[Iterable[int]] = None,
ax: Optional[Axes] = None,
@@ -88,3 +91,18 @@ def plotObservableTrajectories(
ax.set_ylabel('$y(t)$')
ax.legend()
ax.set_title('Observable trajectories')
+
+
+def plot_jacobian(rdata: ReturnDataView):
+ """Plot Jacobian as heatmap."""
+ df = pd.DataFrame(
+ data=rdata.J,
+ index=rdata._swigptr.state_ids_solver,
+ columns=rdata._swigptr.state_ids_solver,
+ )
+ sns.heatmap(df, center=0.0)
+ plt.title("Jacobian")
+
+# backwards compatibility
+plotStateTrajectories = plot_state_trajectories
+plotObservableTrajectories = plot_observable_trajectories
diff --git a/python/sdist/amici/swig.py b/python/sdist/amici/swig.py
index b2ab7c090c..ed1aeb8534 100644
--- a/python/sdist/amici/swig.py
+++ b/python/sdist/amici/swig.py
@@ -133,6 +133,10 @@ def visit_FunctionDef(self, node):
for arg in node.args.args:
if not arg.annotation:
continue
+ if isinstance(arg.annotation, ast.Name):
+ # there is already proper annotation
+ continue
+
arg.annotation = self._new_annot(arg.annotation.value)
return node
diff --git a/python/sdist/amici/swig_wrappers.py b/python/sdist/amici/swig_wrappers.py
index 4b303536ae..cf998970e6 100644
--- a/python/sdist/amici/swig_wrappers.py
+++ b/python/sdist/amici/swig_wrappers.py
@@ -281,3 +281,5 @@ def _ids_and_names_to_rdata(
f"{entity_type.lower()}_{name_or_id.lower()}",
names_or_ids
)
+ rdata.state_ids_solver = model.getStateIdsSolver()
+ rdata.state_names_solver = model.getStateNamesSolver()
diff --git a/python/tests/test_sbml_import.py b/python/tests/test_sbml_import.py
index 8c632c23b5..f70cb773ab 100644
--- a/python/tests/test_sbml_import.py
+++ b/python/tests/test_sbml_import.py
@@ -180,6 +180,11 @@ def test_logging_works(observable_dependent_error_model, caplog):
assert rdata.status != amici.AMICI_SUCCESS
assert "mxstep steps taken" in caplog.text
+@skip_on_valgrind
+def test_model_module_is_set(observable_dependent_error_model):
+ model_module = observable_dependent_error_model
+ assert isinstance(model_module.getModel().module, amici.ModelModule)
+
@pytest.fixture(scope='session')
def model_steadystate_module():
diff --git a/python/tests/test_swig_interface.py b/python/tests/test_swig_interface.py
index c23eaaad00..e7c52d1982 100644
--- a/python/tests/test_swig_interface.py
+++ b/python/tests/test_swig_interface.py
@@ -403,3 +403,34 @@ def test_model_instance_settings_custom_x0(pysb_example_presimulation_module):
assert model2.getInitialStateSensitivities() == sx0
assert settings == amici.get_model_settings(model2)
+
+def test_solver_repr():
+ for solver in (amici.CVodeSolver(), amici.IDASolver()):
+ solver_ptr = amici.SolverPtr(solver.this)
+ for s in (solver, solver_ptr):
+ assert "maxsteps" in str(s)
+ assert "maxsteps" in repr(s)
+ # avoid double delete!!
+ solver_ptr.release()
+
+
+def test_edata_repr():
+ ny = 1
+ nz = 2
+ ne = 3
+ nt = 4
+ edata = amici.ExpData(ny, nz, ne, range(nt))
+ edata_ptr = amici.ExpDataPtr(edata.this)
+ expected_strs = (
+ f'{nt}x{ny} time-resolved datapoints',
+ f'{ne}x{nz} event-resolved datapoints',
+ f'(0/{ny * nt} measurements',
+ f'(0/{nz * ne} measurements'
+ )
+ for e in [edata, edata_ptr]:
+ for expected_str in expected_strs:
+ assert expected_str in str(e)
+ assert expected_str in repr(e)
+ # avoid double delete!!
+ edata_ptr.release()
+
diff --git a/python/tests/valgrind-python.supp b/python/tests/valgrind-python.supp
index 7815e51fa1..dde0c74cf7 100644
--- a/python/tests/valgrind-python.supp
+++ b/python/tests/valgrind-python.supp
@@ -755,3 +755,9 @@
fun:dlopen@@GLIBC_2.34
fun:_PyImport_FindSharedFuncptr
}
+
+{
+ Python dictkeys_get_index
+ Memcheck:Value8
+ fun:dictkeys_get_index
+}
diff --git a/scripts/run-sphinx.sh b/scripts/run-sphinx.sh
index 933b8dc9cf..04716d4b7b 100755
--- a/scripts/run-sphinx.sh
+++ b/scripts/run-sphinx.sh
@@ -7,8 +7,8 @@ AMICI_PATH=$(cd $SCRIPT_PATH/.. && pwd)
python3 -m venv ${AMICI_PATH}/doc-venv --clear
source ${AMICI_PATH}/doc-venv/bin/activate
python -m pip install --upgrade --no-cache-dir pip
-python -m pip install --exists-action=w --no-cache-dir -r ${AMICI_PATH}/documentation/rtd_requirements.txt
-python -m pip install --exists-action=w --no-cache-dir -r ${AMICI_PATH}/documentation/rtd_requirements2.txt
+(cd ${AMICI_PATH}/ && python -m pip install --exists-action=w --no-cache-dir -r documentation/rtd_requirements.txt)
+(cd ${AMICI_PATH}/ && python -m pip install --exists-action=w --no-cache-dir -r documentation/rtd_requirements2.txt)
${AMICI_PATH}/scripts/run-sphinx-hasenv.sh
diff --git a/src/amici.cpp b/src/amici.cpp
index 338937bca1..55060e083a 100644
--- a/src/amici.cpp
+++ b/src/amici.cpp
@@ -51,6 +51,7 @@ std::map simulation_status_to_str_map = {
{AMICI_TOO_MUCH_ACC, "AMICI_TOO_MUCH_ACC"},
{AMICI_ERR_FAILURE, "AMICI_ERR_FAILURE"},
{AMICI_CONV_FAILURE, "AMICI_CONV_FAILURE"},
+ {AMICI_FIRST_RHSFUNC_ERR, "AMICI_FIRST_RHSFUNC_ERR"},
{AMICI_RHSFUNC_FAIL, "AMICI_RHSFUNC_FAIL"},
{AMICI_ILL_INPUT, "AMICI_ILL_INPUT"},
{AMICI_ERROR, "AMICI_ERROR"},
diff --git a/src/exception.cpp b/src/exception.cpp
index 2d1f72ecdc..871c240c9b 100644
--- a/src/exception.cpp
+++ b/src/exception.cpp
@@ -8,43 +8,42 @@
namespace amici {
-AmiException::AmiException()
-{
- storeBacktrace(12);
+AmiException::AmiException(int const first_frame) {
+ storeBacktrace(12, first_frame);
}
-AmiException::AmiException(const char *fmt, ...)
- : AmiException()
-{
+AmiException::AmiException(char const* fmt, ...)
+ : AmiException(4) {
va_list ap;
va_start(ap, fmt);
storeMessage(fmt, ap);
va_end(ap);
}
-const char *AmiException::what() const noexcept {
- return msg_.data();
-}
+char const* AmiException::what() const noexcept { return msg_.data(); }
-const char *AmiException::getBacktrace() const {
- return trace_.data();
-}
+char const* AmiException::getBacktrace() const { return trace_.data(); }
-void AmiException::storeBacktrace(const int nMaxFrames) {
- snprintf(trace_.data(), trace_.size(), "%s",
- backtraceString(nMaxFrames).c_str());
+void AmiException::storeBacktrace(int const nMaxFrames, int const first_frame) {
+ snprintf(
+ trace_.data(), trace_.size(), "%s",
+ backtraceString(nMaxFrames, first_frame).c_str()
+ );
}
-void AmiException::storeMessage(const char *fmt, va_list argptr)
-{
+void AmiException::storeMessage(char const* fmt, va_list argptr) {
vsnprintf(msg_.data(), msg_.size(), fmt, argptr);
}
-CvodeException::CvodeException(const int error_code, const char *function) :
- AmiException("Cvode routine %s failed with error code %i",function,error_code){}
+CvodeException::CvodeException(int const error_code, char const* function)
+ : AmiException(
+ "Cvode routine %s failed with error code %i", function, error_code
+ ) {}
-IDAException::IDAException(const int error_code, const char *function) :
- AmiException("IDA routine %s failed with error code %i",function,error_code){}
+IDAException::IDAException(int const error_code, char const* function)
+ : AmiException(
+ "IDA routine %s failed with error code %i", function, error_code
+ ) {}
IntegrationFailure::IntegrationFailure(int code, realtype t) :
AmiException("AMICI failed to integrate the forward problem"),
@@ -54,13 +53,14 @@ IntegrationFailureB::IntegrationFailureB(int code, realtype t) :
AmiException("AMICI failed to integrate the backward problem"),
error_code(code), time(t) {}
-NewtonFailure::NewtonFailure(int code, const char *function) :
- AmiException("NewtonSolver routine %s failed with error code %i",function,code) {
+NewtonFailure::NewtonFailure(int code, char const* function)
+ : AmiException(
+ "NewtonSolver routine %s failed with error code %i", function, code
+ ) {
error_code = code;
}
-SetupFailure::SetupFailure(const char *fmt, ...)
-{
+SetupFailure::SetupFailure(char const* fmt, ...) {
va_list ap;
va_start(ap, fmt);
storeMessage(fmt, ap);
diff --git a/src/misc.cpp b/src/misc.cpp
index 9c360064ef..bce3fdfddb 100644
--- a/src/misc.cpp
+++ b/src/misc.cpp
@@ -20,7 +20,7 @@
namespace amici {
-void writeSlice(const AmiVector &s, gsl::span b) {
+void writeSlice(AmiVector const& s, gsl::span b) {
writeSlice(s.getVector(), b);
};
@@ -38,10 +38,10 @@ double getUnscaledParameter(double scaledParameter, ParameterScaling scaling)
throw AmiException("Invalid value for ParameterScaling.");
}
-void unscaleParameters(gsl::span bufferScaled,
- gsl::span pscale,
- gsl::span bufferUnscaled)
-{
+void unscaleParameters(
+ gsl::span bufferScaled,
+ gsl::span pscale, gsl::span bufferUnscaled
+) {
Expects(bufferScaled.size() == pscale.size());
Expects(bufferScaled.size() == bufferUnscaled.size());
@@ -51,7 +51,6 @@ void unscaleParameters(gsl::span bufferScaled,
}
}
-
double getScaledParameter(double unscaledParameter, ParameterScaling scaling)
{
switch (scaling) {
@@ -66,11 +65,10 @@ double getScaledParameter(double unscaledParameter, ParameterScaling scaling)
throw AmiException("Invalid value for ParameterScaling.");
}
-
-void scaleParameters(gsl::span bufferUnscaled,
- gsl::span pscale,
- gsl::span bufferScaled)
-{
+void scaleParameters(
+ gsl::span bufferUnscaled,
+ gsl::span pscale, gsl::span bufferScaled
+) {
Expects(bufferScaled.size() == pscale.size());
Expects(bufferScaled.size() == bufferUnscaled.size());
@@ -78,23 +76,21 @@ void scaleParameters(gsl::span bufferUnscaled,
ip < bufferUnscaled.size(); ++ip) {
bufferScaled[ip] = getScaledParameter(bufferUnscaled[ip], pscale[ip]);
}
-
}
-std::string backtraceString(const int maxFrames)
-{
+std::string backtraceString(int const maxFrames, int const first_frame) {
std::ostringstream trace_buf;
#ifdef PLATFORM_WINDOWS
trace_buf << "stacktrace not available on windows platforms\n";
#else
- void *callstack[maxFrames];
+ int const last_frame = first_frame + maxFrames;
+ void* callstack[last_frame];
char buf[1024];
- int nFrames = backtrace(callstack, maxFrames);
+ int nFrames = backtrace(callstack, last_frame);
char **symbols = backtrace_symbols(callstack, nFrames);
- // start at 2 to omit AmiException and storeBacktrace
- for (int i = 2; i < nFrames; i++) {
+ for (int i = first_frame; i < nFrames; i++) {
// call
Dl_info info;
if (dladdr(callstack[i], &info) && info.dli_sname) {
@@ -120,8 +116,8 @@ std::string backtraceString(const int maxFrames)
}
free(symbols);
- if (nFrames == maxFrames)
- trace_buf << "[truncated]\n";
+ if (nFrames == last_frame)
+ trace_buf << "[possibly truncated]\n";
#endif
return trace_buf.str();
}
@@ -160,7 +156,7 @@ std::string regexErrorToString(std::regex_constants::error_type err_type)
}
}
-std::string printfToString(const char *fmt, va_list ap) {
+std::string printfToString(char const* fmt, va_list ap) {
// Get size of string
va_list ap_count;
va_copy(ap_count, ap);
diff --git a/src/model.cpp b/src/model.cpp
index df71ae3b8f..db36040167 100644
--- a/src/model.cpp
+++ b/src/model.cpp
@@ -305,6 +305,8 @@ void Model::initializeStates(AmiVector &x) {
fx_solver(x0_solver.data(), x0data_.data());
std::copy(x0_solver.cbegin(), x0_solver.cend(), x.data());
}
+
+ checkFinite(x.getVector(), ModelQuantity::x0);
}
void Model::initializeStateSensitivities(AmiVectorArray &sx,
@@ -1663,10 +1665,7 @@ void Model::fx0(AmiVector &x) {
state_.unscaledParameters.data(),
state_.fixedParameters.data());
- if (always_check_finite_) {
- checkFinite(derived_state_.x_rdata_, ModelQuantity::x0_rdata);
- checkFinite(x.getVector(), ModelQuantity::x0);
- }
+ checkFinite(derived_state_.x_rdata_, ModelQuantity::x0_rdata);
}
void Model::fx0_fixedParameters(AmiVector &x) {
diff --git a/src/returndata_matlab.cpp b/src/returndata_matlab.cpp
index 297c6d68cf..9599be2673 100644
--- a/src/returndata_matlab.cpp
+++ b/src/returndata_matlab.cpp
@@ -216,7 +216,7 @@ void writeMatlabField1(
double *array = initAndAttachArray(matlabStruct, fieldName, dim);
auto data_ptr = fieldData.data();
- for(int i = 0; i < dim0; i++)
+ for (mwSize i = 0; i < dim0; i++)
array[i] = static_cast(data_ptr[i]);
}
diff --git a/swig/edata.i b/swig/edata.i
index ec6fe7d551..310869768c 100644
--- a/swig/edata.i
+++ b/swig/edata.i
@@ -8,5 +8,81 @@ using namespace amici;
%ignore ConditionContext;
+// ExpData.__repr__
+%pythoncode %{
+def _edata_repr(self: "ExpData"):
+ n_data_y = sum(
+ self.isSetObservedData(it, iy)
+ for it in range(self.nt()) for
+ iy in range(self.nytrue())
+ )
+ n_sigma_y = sum(
+ self.isSetObservedDataStdDev(it, iy)
+ for it in range(self.nt())
+ for iy in range(self.nytrue())
+ )
+ n_data_z = sum(
+ self.isSetObservedEvents(ie, iz)
+ for ie in range(self.nmaxevent())
+ for iz in range(self.nztrue())
+ )
+ n_sigma_z = sum(
+ self.isSetObservedEventsStdDev(ie, iz)
+ for ie in range(self.nmaxevent())
+ for iz in range(self.nztrue())
+ )
+
+ custom_simulation_settings = []
+ if self.pscale:
+ custom_simulation_settings.append(f"parameter scales")
+ if self.fixedParameters:
+ custom_simulation_settings.append(f"constants")
+ if self.fixedParametersPreequilibration:
+ custom_simulation_settings.append(f"pre-equilibration condition")
+ if self.t_presim:
+ tmp = f"pre-simulation condition (t={self.t_presim})"
+ if self.fixedParametersPresimulation:
+ tmp += " with custom constants"
+ custom_simulation_settings.append(tmp)
+ if self.reinitializeFixedParameterInitialStates and self.reinitialization_state_idxs_sim:
+ custom_simulation_settings.append(f"{len(self.reinitialization_state_idxs_sim)} reinitialized states (simulation)")
+ if self.reinitializeFixedParameterInitialStates and self.reinitialization_state_idxs_presim:
+ custom_simulation_settings.append(f"{len(self.reinitialization_state_idxs_presim)} reinitialized states (presimulation)")
+ if self.parameters:
+ custom_simulation_settings.append(f"parameters")
+ if self.x0:
+ custom_simulation_settings.append(f"initial states")
+ if self.sx0:
+ custom_simulation_settings.append(f"initial state sensitivities")
+
+ if custom_simulation_settings:
+ custom_simulation_settings = " with custom " + ", ".join(custom_simulation_settings)
+ else:
+ custom_simulation_settings = " without custom settings"
+
+ return "\n".join([
+ self.this.__repr__()[:-1],
+ f" condition {id} starting at t={self.tstart_}" + custom_simulation_settings,
+ f" {self.nt()}x{self.nytrue()} time-resolved datapoints",
+ f" ({n_data_y}/{self.nt()*self.nytrue()} measurements & {n_sigma_y}/{self.nt()*self.nytrue()} sigmas set)",
+ f" {self.nmaxevent()}x{self.nztrue()} event-resolved datapoints",
+ f" ({n_data_z}/{self.nmaxevent()*self.nztrue()} measurements & {n_sigma_z}/{self.nmaxevent()*self.nztrue()} sigmas set)",
+ ">"
+ ])
+%}
+%extend amici::ExpData {
+%pythoncode %{
+def __repr__(self):
+ return _edata_repr(self)
+%}
+};
+%extend std::unique_ptr {
+%pythoncode %{
+def __repr__(self):
+ return _edata_repr(self)
+%}
+};
+
+
// Process symbols in header
%include "amici/edata.h"
diff --git a/swig/modelname.template.i b/swig/modelname.template.i
index 3425d78a17..33804a1800 100644
--- a/swig/modelname.template.i
+++ b/swig/modelname.template.i
@@ -10,5 +10,12 @@ using namespace amici;
%}
+// Make model module accessible from the model
+%feature("pythonappend") amici::generic_model::getModel %{
+ import sys
+ val.module = sys.modules['.'.join(__name__.split('.')[:-1])]
+%}
+
+
// Process symbols in header
%include "wrapfunctions.h"
diff --git a/swig/solver.i b/swig/solver.i
index 853edae9bd..319e654530 100644
--- a/swig/solver.i
+++ b/swig/solver.i
@@ -40,6 +40,67 @@ using namespace amici;
%ignore getRootInfo;
%ignore updateAndReinitStatesAndSensitivities;
+// Solver.__repr__
+%pythoncode %{
+def _solver_repr(self: "Solver"):
+ return "\n".join([
+ self.this.__repr__()[:-1],
+ " reporting_mode: "
+ f"{RDataReporting(self.getReturnDataReportingMode())!r}",
+ f" sens_meth: {SensitivityMethod(self.getSensitivityMethod())!r}",
+ f" sens_order: {SensitivityOrder(self.getSensitivityOrder())!r}",
+ f" sens_meth_preeq: {SensitivityMethod(self.getSensitivityMethodPreequilibration())!r}",
+ f" maxsteps: {self.getMaxSteps()}",
+ f" maxtime: {self.getMaxTime()}s",
+ f" abs_tol: {self.getAbsoluteTolerance()}",
+ f" rel_tol: {self.getRelativeTolerance()}",
+ f" abs_tol_b: {self.getAbsoluteToleranceB()}",
+ f" rel_tol_b: {self.getRelativeToleranceB()}",
+ f" abs_tol_fsa: {self.getAbsoluteToleranceFSA()}",
+ f" rel_tol_fsa: {self.getRelativeToleranceFSA()}",
+ f" abs_tol_quad: {self.getAbsoluteToleranceQuadratures()}",
+ f" rel_tol_quad: {self.getRelativeToleranceQuadratures()}",
+ f" abs_tol_ss: {self.getAbsoluteToleranceSteadyState()}",
+ f" rel_tol_ss: {self.getRelativeToleranceSteadyState()}",
+ f" abs_tol_sss: {self.getAbsoluteToleranceSteadyStateSensi()}",
+ f" rel_tol_sss: {self.getRelativeToleranceSteadyStateSensi()}",
+ f" int_sens_meth: {InternalSensitivityMethod(self.getInternalSensitivityMethod())!r}",
+ f" int_type: {InterpolationType(self.getInterpolationType())!r}",
+ f" linsol: {LinearSolver(self.getLinearSolver())!r}",
+ f" lmm: {LinearMultistepMethod(self.getLinearMultistepMethod())!r}",
+ f" newton_damp_mode: {NewtonDampingFactorMode(self.getNewtonDampingFactorMode())!r}",
+ f" newton_damp_lb: {self.getNewtonDampingFactorLowerBound()}",
+ f" newton_maxsteps: {self.getNewtonMaxSteps()}",
+ f" newton_ss_check: {self.getNewtonStepSteadyStateCheck()}",
+ f" sens_ss_check: {self.getSensiSteadyStateCheck()}",
+ f" interpolation_type: {InterpolationType(self.getInterpolationType())!r}",
+ f" ism: {InternalSensitivityMethod(self.getInternalSensitivityMethod())!r}",
+ f" nlsol_iter: {NonlinearSolverIteration(self.getNonlinearSolverIteration())!r}",
+ f" stability_limit: {self.getStabilityLimitFlag()}",
+ f" state_ordering: {self.getStateOrdering()}",
+ ">"
+ ])
+%}
+%extend amici::CVodeSolver {
+%pythoncode %{
+def __repr__(self):
+ return _solver_repr(self)
+%}
+};
+%extend amici::IDASolver {
+%pythoncode %{
+def __repr__(self):
+ return _solver_repr(self)
+%}
+};
+
+%extend std::unique_ptr {
+%pythoncode %{
+def __repr__(self):
+ return _solver_repr(self)
+%}
+};
+
%newobject amici::Solver::clone;
// Process symbols in header
diff --git a/tests/benchmark-models/benchmark_models.yaml b/tests/benchmark-models/benchmark_models.yaml
index d9ce6d0dd9..42d9f4a71d 100644
--- a/tests/benchmark-models/benchmark_models.yaml
+++ b/tests/benchmark-models/benchmark_models.yaml
@@ -1,15 +1,15 @@
Bachmann_MSB2011:
llh: 418.40573341425295
t_sim: 0.05
- t_fwd: 3.0
- t_adj: 4.5
+ t_fwd: 3.5
+ t_adj: 5.0
note: benchmark collection reference value matches up to sign when applying log10-correction +sum(log(meas*log(10)) / 2
Beer_MolBioSystems2014:
llh: 58622.9145631413
t_sim: 0.05
t_fwd: 0.5
- t_adj: 8.0
+ t_adj: 8.5
note: benchmark collection reference parameters do not match petab, but reference llh has been confirmed for parameters reported there up to sign
Boehm_JProteomeRes2014:
@@ -61,7 +61,7 @@ Fiedler_BMC2016:
llh: 58.58390161681
t_sim: 0.005
t_fwd: 0.05
- t_adj: 0.1
+ t_adj: 0.15
Fujita_SciSignal2010:
llh: 53.08377736998929
@@ -80,7 +80,7 @@ Lucarelli_CellSystems2018:
llh: -1681.6059879426584
t_sim: 0.05
t_fwd: 2.5
- t_adj: 2.0
+ t_adj: 2.5
Schwen_PONE2014:
llh: -943.9992988598723
diff --git a/tests/performance/test.py b/tests/performance/test.py
index 9dde939f5f..03a2309c77 100755
--- a/tests/performance/test.py
+++ b/tests/performance/test.py
@@ -4,6 +4,7 @@
import shutil
import subprocess
import sys
+import logging
from pathlib import Path
import amici
@@ -47,6 +48,8 @@ def run_import(model_name, model_dir: Path):
pp = petab.Problem.from_yaml(git_dir / 'FroehlichKes2018' / 'PEtab'
/ 'FroehlichKes2018.yaml')
petab.lint_problem(pp)
+ amici.ode_export.logger.setLevel(logging.DEBUG)
+ amici.sbml_import.logger.setLevel(logging.DEBUG)
import_model(model_name=model_name,
model_output_dir=model_dir,
sbml_model=pp.sbml_model,
diff --git a/version.txt b/version.txt
index a551051694..04a373efe6 100644
--- a/version.txt
+++ b/version.txt
@@ -1 +1 @@
-0.15.0
+0.16.0